Prediction of enzyme function based on 3D templates of evolutionarily important amino acids
- PMID: 18190718
- PMCID: PMC2219985
- DOI: 10.1186/1471-2105-9-17
Prediction of enzyme function based on 3D templates of evolutionarily important amino acids
Abstract
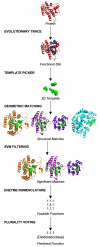
Background: Structural genomics projects such as the Protein Structure Initiative (PSI) yield many new structures, but often these have no known molecular functions. One approach to recover this information is to use 3D templates - structure-function motifs that consist of a few functionally critical amino acids and may suggest functional similarity when geometrically matched to other structures. Since experimentally determined functional sites are not common enough to define 3D templates on a large scale, this work tests a computational strategy to select relevant residues for 3D templates.
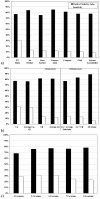
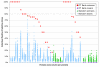
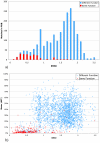
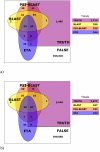
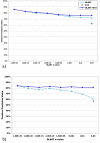
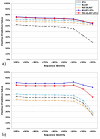
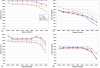
Results: Based on evolutionary information and heuristics, an Evolutionary Trace Annotation (ETA) pipeline built templates for 98 enzymes, half taken from the PSI, and sought matches in a non-redundant structure database. On average each template matched 2.7 distinct proteins, of which 2.0 share the first three Enzyme Commission digits as the template's enzyme of origin. In many cases (61%) a single most likely function could be predicted as the annotation with the most matches, and in these cases such a plurality vote identified the correct function with 87% accuracy. ETA was also found to be complementary to sequence homology-based annotations. When matches are required to both geometrically match the 3D template and to be sequence homologs found by BLAST or PSI-BLAST, the annotation accuracy is greater than either method alone, especially in the region of lower sequence identity where homology-based annotations are least reliable.
Conclusion: These data suggest that knowledge of evolutionarily important residues improves functional annotation among distant enzyme homologs. Since, unlike other 3D template approaches, the ETA method bypasses the need for experimental knowledge of the catalytic mechanism, it should prove a useful, large scale, and general adjunct to combine with other methods to decipher protein function in the structural proteome.
Figures

Similar articles
-
Evolutionary trace annotation of protein function in the structural proteome.J Mol Biol. 2010 Mar 12;396(5):1451-73. doi: 10.1016/j.jmb.2009.12.037. Epub 2009 Dec 28. J Mol Biol. 2010. PMID: 20036248 Free PMC article.
-
Evolutionary Trace Annotation Server: automated enzyme function prediction in protein structures using 3D templates.Bioinformatics. 2009 Jun 1;25(11):1426-7. doi: 10.1093/bioinformatics/btp160. Epub 2009 Mar 23. Bioinformatics. 2009. PMID: 19307237 Free PMC article.
-
Recurrent use of evolutionary importance for functional annotation of proteins based on local structural similarity.Protein Sci. 2006 Jun;15(6):1530-6. doi: 10.1110/ps.062152706. Epub 2006 May 2. Protein Sci. 2006. PMID: 16672239 Free PMC article.
-
Template-based prediction of protein function.Curr Opin Struct Biol. 2015 Jun;32:33-8. doi: 10.1016/j.sbi.2015.01.007. Epub 2015 Feb 10. Curr Opin Struct Biol. 2015. PMID: 25678152 Free PMC article. Review.
-
Proteins: sequence to structure and function--current status.Curr Protein Pept Sci. 2010 Nov;11(7):498-514. doi: 10.2174/138920310794109094. Curr Protein Pept Sci. 2010. PMID: 20887265 Review.
Cited by
-
Function prediction from networks of local evolutionary similarity in protein structure.BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S6. doi: 10.1186/1471-2105-14-S3-S6. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514548 Free PMC article.
-
Structure prediction of partial-length protein sequences.Int J Mol Sci. 2013 Jul 17;14(7):14892-907. doi: 10.3390/ijms140714892. Int J Mol Sci. 2013. PMID: 23867606 Free PMC article.
-
Prediction of detailed enzyme functions and identification of specificity determining residues by random forests.PLoS One. 2014 Jan 8;9(1):e84623. doi: 10.1371/journal.pone.0084623. eCollection 2014. PLoS One. 2014. PMID: 24416252 Free PMC article.
-
Sequence and structure continuity of evolutionary importance improves protein functional site discovery and annotation.Protein Sci. 2010 Jul;19(7):1296-311. doi: 10.1002/pro.406. Protein Sci. 2010. PMID: 20506260 Free PMC article.
-
Capturing the geometry, function, and evolution of enzymes with 3D templates.Protein Sci. 2022 Jul;31(7):e4363. doi: 10.1002/pro.4363. Protein Sci. 2022. PMID: 35762726 Free PMC article. Review.
References
-
- Brenner SE. A tour of structural genomics. Nat Rev Genet. 2001;2:801–809. - PubMed
-
- Burley SK. An overview of structural genomics. Nat Struct Biol. 2000;7 Suppl:932–934. - PubMed
-
- Leulliot N, Tresaugues L, Bremang M, Sorel I, Ulryck N, Graille M, Aboulfath I, Poupon A, Liger D, Quevillon-Cheruel S, Janin J, van Tilbeurgh H. High-throughput crystal-optimization strategies in the South Paris Yeast Structural Genomics Project: one size fits all? Acta Crystallogr D Biol Crystallogr. 2005;61:664–670. - PubMed
-
- Kuznetsova E, Proudfoot M, Sanders SA, Reinking J, Savchenko A, Arrowsmith CH, Edwards AM, Yakunin AF. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol Rev. 2005;29:263–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

