DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication
- PMID: 18158334
- PMCID: PMC2373486
- DOI: 10.1083/jcb.200708106
DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication
Erratum in
- J Cell Biol. 2008 Feb 25;180(4):843
Abstract
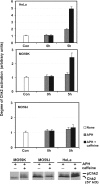
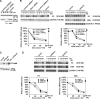
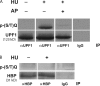
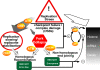
DNA and histone synthesis are coupled and ongoing replication is required to maintain histone gene expression. Here, we expose S phase-arrested cells to the kinase inhibitors caffeine and LY294002. This uncouples DNA replication from histone messenger RNA (mRNA) abundance, altering the efficiency of replication stress-induced histone mRNA down-regulation. Interference with caffeine-sensitive checkpoint kinases ataxia telangiectasia and Rad3 related (ATR)/ataxia telangiectasia mutated (ATM) does not affect histone mRNA down- regulation, which indicates that ATR/ATM alone cannot account for such coupling. LY294002 potentiates caffeine's ability to uncouple histone mRNA stabilization from replication only in cells containing functional DNA-activated protein kinase (DNA-PK), which indicates that DNA-PK is the target of LY294002. DNA-PK is activated during replication stress and DNA-PK signaling is enhanced when ATR/ATM signaling is abrogated. Histone mRNA decay does not require Chk1/Chk2. Replication stress induces phosphorylation of UPF1 but not hairpin[corrected]-binding protein/stem-loop binding protein at S/TQ sites, which are preferred substrate recognition motifs of phosphatidylinositol 3-kinase-like kinases, which indicates that histone mRNA stability may be directly controlled by ATR/ATM- and DNA-PK-mediated phosphorylation of UPF1.
Figures





Similar articles
-
Chk1- and claspin-dependent but ATR/ATM- and Rad17-independent DNA replication checkpoint response in HeLa cells.Cancer Res. 2006 Sep 1;66(17):8672-9. doi: 10.1158/0008-5472.CAN-05-4443. Cancer Res. 2006. PMID: 16951182
-
Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1.Nat Struct Mol Biol. 2005 Sep;12(9):794-800. doi: 10.1038/nsmb972. Epub 2005 Aug 7. Nat Struct Mol Biol. 2005. PMID: 16086026
-
Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase.Cancer Res. 2001 Dec 1;61(23):8554-63. Cancer Res. 2001. PMID: 11731442
-
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer.Adv Cancer Res. 2010;108:73-112. doi: 10.1016/B978-0-12-380888-2.00003-0. Adv Cancer Res. 2010. PMID: 21034966 Review.
-
The ATR-independent DNA replication checkpoint.Cell Cycle. 2003 May-Jun;2(3):188-9. Cell Cycle. 2003. PMID: 12734420 Review. No abstract available.
Cited by
-
Tracing the molecular basis of transcriptional dynamics in noisy data by using an experiment-based mathematical model.Nucleic Acids Res. 2015 Jan;43(1):153-61. doi: 10.1093/nar/gku1272. Epub 2014 Dec 3. Nucleic Acids Res. 2015. PMID: 25477385 Free PMC article.
-
Signaling pathways that control mRNA turnover.Cell Signal. 2013 Aug;25(8):1699-710. doi: 10.1016/j.cellsig.2013.03.026. Epub 2013 Apr 16. Cell Signal. 2013. PMID: 23602935 Free PMC article. Review.
-
More forks on the road to replication stress recovery.J Mol Cell Biol. 2011 Feb;3(1):4-12. doi: 10.1093/jmcb/mjq049. J Mol Cell Biol. 2011. PMID: 21278446 Free PMC article. Review.
-
The multiple lives of NMD factors: balancing roles in gene and genome regulation.Nat Rev Genet. 2008 Sep;9(9):699-712. doi: 10.1038/nrg2402. Nat Rev Genet. 2008. PMID: 18679436 Free PMC article. Review.
-
Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation.Proc Natl Acad Sci U S A. 2015 Mar 31;112(13):E1540-9. doi: 10.1073/pnas.1409612112. Epub 2015 Mar 16. Proc Natl Acad Sci U S A. 2015. PMID: 25775514 Free PMC article.
References
-
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177–2196. - PubMed
-
- Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235–1245. - PubMed
-
- Azzalin, C.M., and J. Lingner. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16:433–439. - PubMed
-
- Baumbach, L.L., G.S. Stein, and J.L. Stein. 1987. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 26:6178–6187. - PubMed
-
- Bhattacharyya, N.P., A. Ganesh, G. Phear, B. Richards, A. Skandalis, and M. Meuth. 1995. Molecular analysis of mutations in mutator colorectal carcinoma cell lines. Hum. Mol. Genet. 4:2057–2064. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

