Identification of transient hub proteins and the possible structural basis for their multiple interactions
- PMID: 18156468
- PMCID: PMC2144588
- DOI: 10.1110/ps.073196308
Identification of transient hub proteins and the possible structural basis for their multiple interactions
Abstract
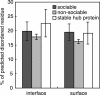
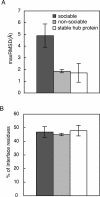
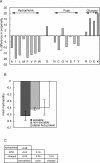
Proteins that can interact with multiple partners play central roles in the network of protein-protein interactions. They are called hub proteins, and recently it was suggested that an abundance of intrinsically disordered regions on their surfaces facilitates their binding to multiple partners. However, in those studies, the hub proteins were identified as proteins with multiple partners, regardless of whether the interactions were transient or permanent. As a result, a certain number of hub proteins are subunits of stable multi-subunit proteins, such as supramolecules. It is well known that stable complexes and transient complexes have different structural features, and thus the statistics based on the current definition of hub proteins will hide the true nature of hub proteins. Therefore, in this paper, we first describe a new approach to identify proteins with multiple partners dynamically, using the Protein Data Bank, and then we performed statistical analyses of the structural features of these proteins. We refer to the proteins as transient hub proteins or sociable proteins, to clarify the difference with hub proteins. As a result, we found that the main difference between sociable and nonsociable proteins is not the abundance of disordered regions, in contrast to the previous studies, but rather the structural flexibility of the entire protein. We also found greater predominance of charged and polar residues in sociable proteins than previously reported.
Figures

Similar articles
-
Distinct roles of overlapping and non-overlapping regions of hub protein interfaces in recognition of multiple partners.J Mol Biol. 2011 Aug 19;411(3):713-27. doi: 10.1016/j.jmb.2011.06.027. Epub 2011 Jun 22. J Mol Biol. 2011. PMID: 21723293
-
Evolutionary constraints on hub and non-hub proteins in human protein interaction network: insight from protein connectivity and intrinsic disorder.Gene. 2009 Apr 1;434(1-2):50-5. doi: 10.1016/j.gene.2008.12.013. Epub 2008 Dec 29. Gene. 2009. PMID: 19185053
-
Analysis of molecular recognition features (MoRFs).J Mol Biol. 2006 Oct 6;362(5):1043-59. doi: 10.1016/j.jmb.2006.07.087. Epub 2006 Aug 4. J Mol Biol. 2006. PMID: 16935303
-
The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins.Phys Biol. 2011 Jun;8(3):035003. doi: 10.1088/1478-3975/8/3/035003. Epub 2011 May 13. Phys Biol. 2011. PMID: 21572179 Review.
-
Structural polymorphism and multifunctionality of myelin basic protein.Biochemistry. 2009 Sep 1;48(34):8094-104. doi: 10.1021/bi901005f. Biochemistry. 2009. PMID: 19642704 Review.
Cited by
-
Assessing the utility of gene co-expression stability in combination with correlation in the analysis of protein-protein interaction networks.BMC Genomics. 2011 Nov 30;12 Suppl 3(Suppl 3):S19. doi: 10.1186/1471-2164-12-S3-S19. Epub 2011 Nov 30. BMC Genomics. 2011. PMID: 22369639 Free PMC article.
-
Reciprocal Perspective for Improved Protein-Protein Interaction Prediction.Sci Rep. 2018 Aug 3;8(1):11694. doi: 10.1038/s41598-018-30044-1. Sci Rep. 2018. PMID: 30076341 Free PMC article.
-
Intrinsic disorder in scaffold proteins: getting more from less.Prog Biophys Mol Biol. 2008 Sep;98(1):85-106. doi: 10.1016/j.pbiomolbio.2008.05.007. Epub 2008 Jun 20. Prog Biophys Mol Biol. 2008. PMID: 18619997 Free PMC article. Review.
-
Protein-protein interaction network prediction by using rigid-body docking tools: application to bacterial chemotaxis.Protein Pept Lett. 2014;21(8):790-8. doi: 10.2174/09298665113209990066. Protein Pept Lett. 2014. PMID: 23855669 Free PMC article.
-
PiSite: a database of protein interaction sites using multiple binding states in the PDB.Nucleic Acids Res. 2009 Jan;37(Database issue):D360-4. doi: 10.1093/nar/gkn659. Epub 2008 Oct 4. Nucleic Acids Res. 2009. PMID: 18836195 Free PMC article.
References
-
- Bracken, C., Iakoucheva, L.M., Romero, P.R., Dunker, A.K. Combining prediction, computation and experiment for the characterization of protein disorder. Curr. Opin. Struct. Biol. 2004;14:570–576. - PubMed
-
- Dunker, A.K., Cortese, M.S., Romero, P., Iakoucheva, L.M., Uversky, V.N. Flexible nets—The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. - PubMed
-
- Dyson, H.J., Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. - PubMed
-
- Eisenhaber, F., Argos, P. Improved strategy in analytic surface calculation for molecular systems: Handling of singularities and computational efficiency. J. Comput. Chem. 1993;14:1272–1280.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

