Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant
- PMID: 18156258
- PMCID: PMC2258657
- DOI: 10.1128/JB.01463-07
Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant
Abstract
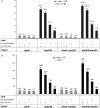
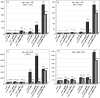
The dnaX36(TS) mutant of Escherichia coli confers a distinct mutator phenotype characterized by enhancement of transversion base substitutions and certain (-1) frameshift mutations. Here, we have further investigated the possible mechanism(s) underlying this mutator effect, focusing in particular on the role of the various E. coli DNA polymerases. The dnaX gene encodes the tau subunit of DNA polymerase III (Pol III) holoenzyme, the enzyme responsible for replication of the bacterial chromosome. The dnaX36 defect resides in the C-terminal domain V of tau, essential for interaction of tau with the alpha (polymerase) subunit, suggesting that the mutator phenotype is caused by an impaired or altered alpha-tau interaction. We previously proposed that the mutator activity results from aberrant processing of terminal mismatches created by Pol III insertion errors. The present results, including lack of interaction of dnaX36 with mutM, mutY, and recA defects, support our assumption that dnaX36-mediated mutations originate as errors of replication rather than DNA damage-related events. Second, an important role is described for DNA Pol II and Pol IV in preventing and producing, respectively, the mutations. In the system used, a high fraction of the mutations is dependent on the action of Pol IV in a (dinB) gene dosage-dependent manner. However, an even larger but opposing role is deduced for Pol II, revealing Pol II to be a major editor of Pol III mediated replication errors. Overall, the results provide insight into the interplay of the various DNA polymerases, and of tau subunit, in securing a high fidelity of replication.
Figures
Similar articles
-
dnaX36 Mutator of Escherichia coli: effects of the {tau} subunit of the DNA polymerase III holoenzyme on chromosomal DNA replication fidelity.J Bacteriol. 2011 Jan;193(1):296-300. doi: 10.1128/JB.01191-10. Epub 2010 Oct 29. J Bacteriol. 2011. PMID: 21036999 Free PMC article.
-
Mutator mutants of Escherichia coli carrying a defect in the DNA polymerase III tau subunit.Mol Microbiol. 2006 Feb;59(4):1149-61. doi: 10.1111/j.1365-2958.2005.05011.x. Mol Microbiol. 2006. PMID: 16430690
-
DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli.Mol Microbiol. 2005 Oct;58(1):61-70. doi: 10.1111/j.1365-2958.2005.04805.x. Mol Microbiol. 2005. PMID: 16164549
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
Properties and functions of Escherichia coli: Pol IV and Pol V.Adv Protein Chem. 2004;69:229-64. doi: 10.1016/S0065-3233(04)69008-5. Adv Protein Chem. 2004. PMID: 15588845 Review.
Cited by
-
Limits and patterns of cytomegalovirus genomic diversity in humans.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):E4120-8. doi: 10.1073/pnas.1501880112. Epub 2015 Jul 6. Proc Natl Acad Sci U S A. 2015. PMID: 26150505 Free PMC article.
-
Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity.Mol Microbiol. 2009 Dec;74(5):1114-27. doi: 10.1111/j.1365-2958.2009.06921.x. Epub 2009 Oct 19. Mol Microbiol. 2009. PMID: 19843230 Free PMC article.
-
Essential roles for imuA'- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis.Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13093-8. doi: 10.1073/pnas.1002614107. Epub 2010 Jul 6. Proc Natl Acad Sci U S A. 2010. PMID: 20615954 Free PMC article.
-
Effect of dNTP pool alterations on fidelity of leading and lagging strand DNA replication in E. coli.Mutat Res. 2014 Jan;759:22-8. doi: 10.1016/j.mrfmmm.2013.11.003. Epub 2013 Nov 22. Mutat Res. 2014. PMID: 24269257 Free PMC article.
-
DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair.FEMS Microbiol Rev. 2012 Nov;36(6):1105-21. doi: 10.1111/j.1574-6976.2012.00338.x. Epub 2012 Apr 5. FEMS Microbiol Rev. 2012. PMID: 22404288 Free PMC article. Review.
References
-
- Banach-Orlowska, M., I. J. Fijalkowska, R. M. Schaaper, and P. Jonczyk. 2005. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol. Microbiol. 5861-70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

