Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases
- PMID: 18154319
- PMCID: PMC2424258
- DOI: 10.1021/bi701619u
Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases
Abstract
Under conditions of oxidative stress, the formamidopyrimidine lesions (FapyG and FapyA) are formed in competition with the corresponding 8-oxopurines (OG and OA) from a common intermediate. In order to reveal features of the repair of these lesions, and the potential contribution of repair in mitigating or exacerbating the mutagenic properties of Fapy lesions, their excision by three glycosylases, Fpg, hOGG1 and Ntg1, was examined in various base pair contexts under single-turnover conditions. FapyG was removed at least as efficiently as OG by all three glycosylases. In addition, the rates of removal of FapyG by Fpg and hOGG1 were influenced by their base pair partner, with preference for removal when base paired with the correct Watson-Crick partner C. With the FapyA lesion, Fpg and Ntg1 catalyze its removal more readily than OG opposite all four natural bases. In contrast, the removal of FapyA by hOGG1 was not as robust as FapyG or OG, and was only significant when the lesion was paired with C. The discrimination by the various glycosylases with respect to the opposing base was highly dependent on the identity of the lesion. OG induced the greatest selectivity against its removal when part of a promutagenic base pair. The superb activity of the various OG glycosylases toward removal of FapyG and FapyA in vitro suggests that these enzymes may act upon these oxidized lesions in vivo. The differences in the activity of the various glycosylases for removal of FapyG and FapyA compared to OG in nonmutagenic versus promutagenic base pair contexts may serve to alter the mutagenic profiles of these lesions in vivo.
Figures
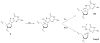


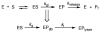
Similar articles
-
Repair of DNA containing Fapy.dG and its beta-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY.Biochemistry. 2003 Aug 19;42(32):9755-60. doi: 10.1021/bi034844h. Biochemistry. 2003. PMID: 12911318
-
Probing the requirements for recognition and catalysis in Fpg and MutY with nonpolar adenine isosteres.J Am Chem Soc. 2003 Dec 31;125(52):16235-42. doi: 10.1021/ja0374426. J Am Chem Soc. 2003. PMID: 14692765
-
Structural Insight into the Discrimination between 8-Oxoguanine Glycosidic Conformers by DNA Repair Enzymes: A Molecular Dynamics Study of Human Oxoguanine Glycosylase 1 and Formamidopyrimidine-DNA Glycosylase.Biochemistry. 2018 Feb 20;57(7):1144-1154. doi: 10.1021/acs.biochem.7b01292. Epub 2018 Feb 6. Biochemistry. 2018. PMID: 29320630
-
Imidazole ring-opened DNA purines and their biological significance.J Biochem Mol Biol. 2003 Jan 31;36(1):12-9. doi: 10.5483/bmbrep.2003.36.1.012. J Biochem Mol Biol. 2003. PMID: 12542970 Review.
-
Repair of oxidative DNA damage: mechanisms and functions.Cell Biochem Biophys. 2001;35(2):141-70. doi: 10.1385/CBB:35:2:141. Cell Biochem Biophys. 2001. PMID: 11892789 Review.
Cited by
-
Structural Dynamics of a Common Mutagenic Oxidative DNA Lesion in Duplex DNA and during DNA Replication.J Am Chem Soc. 2022 May 11;144(18):8054-8065. doi: 10.1021/jacs.2c00193. Epub 2022 May 2. J Am Chem Soc. 2022. PMID: 35499923 Free PMC article.
-
Inflammation-induced DNA damage, mutations and cancer.DNA Repair (Amst). 2019 Nov;83:102673. doi: 10.1016/j.dnarep.2019.102673. Epub 2019 Jul 25. DNA Repair (Amst). 2019. PMID: 31387777 Free PMC article. Review.
-
Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts.Biochemistry. 2010 Mar 2;49(8):1658-66. doi: 10.1021/bi901852q. Biochemistry. 2010. PMID: 20099873 Free PMC article.
-
Fanconi anemia proteins and endogenous stresses.Mutat Res. 2009 Jul 31;668(1-2):42-53. doi: 10.1016/j.mrfmmm.2009.03.013. Mutat Res. 2009. PMID: 19774700 Free PMC article.
-
Synthesis and characterization of oligonucleotides containing a nitrogen mustard formamidopyrimidine monoadduct of deoxyguanosine.Chem Res Toxicol. 2014 Sep 15;27(9):1610-8. doi: 10.1021/tx5002354. Epub 2014 Aug 28. Chem Res Toxicol. 2014. PMID: 25136769 Free PMC article.
References
-
- Neeley WL, Essigmann JM. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem Res Toxicol. 2006;19:491–505. - PubMed
-
- Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. - PubMed
-
- Klaunig JE, Kamendulis LM. The Role of Oxidative Stress in Carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–67. - PubMed
-
- Cheadle JP, Sampson JR. MUTYH-associated polyposis-From defect in base excision repair to clinical genetic testing. DNA Repair. 2006;6:274–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

