HSP105 interacts with GRP78 and GSK3 and promotes ER stress-induced caspase-3 activation
- PMID: 18083346
- PMCID: PMC2212615
- DOI: 10.1016/j.cellsig.2007.10.032
HSP105 interacts with GRP78 and GSK3 and promotes ER stress-induced caspase-3 activation
Abstract
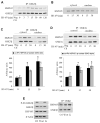
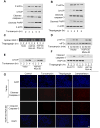
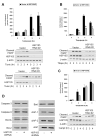
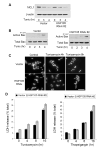
Stress of the endoplasmic reticulum (ER stress) is caused by the accumulation of misfolded proteins, which occurs in many neurodegenerative diseases. ER stress can lead to adaptive responses or apoptosis, both of which follow activation of the unfolded protein response (UPR). Heat shock proteins (HSP) support the folding and function of many proteins, and are important components of the ER stress response, but little is known about the role of one of the major large HSPs, HSP105. We identified several new partners of HSP105, including glycogen synthase kinase-3 (GSK3), a promoter of ER stress-induced apoptosis, and GRP78, a key component of the UPR. Knockdown of HSP105 did not alter UPR signaling after ER stress, but blocked caspase-3 activation after ER stress. In contrast, caspase-3 activation induced by genotoxic stress was unaffected by knockdown of HSP105, suggesting ER stress-specificity in the apoptotic action of HSP105. However, knockdown of HSP105 did not alter cell survival after ER stress, but instead diverted signaling to a caspase-3-independent cell death pathway, indicating that HSP105 is necessary for apoptotic signaling after UPR activation by ER stress. Thus, HSP105 appears to chaperone the responses to ER stress through its interactions with GRP78 and GSK3, and without HSP105 cell death following ER stress proceeds by a non-caspase-3-dependent process.
Figures


Similar articles
-
Activation of PERK signaling attenuates Abeta-mediated ER stress.PLoS One. 2010 May 5;5(5):e10489. doi: 10.1371/journal.pone.0010489. PLoS One. 2010. PMID: 20463975 Free PMC article.
-
Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78.FEBS Lett. 2002 Mar 13;514(2-3):122-8. doi: 10.1016/s0014-5793(02)02289-5. FEBS Lett. 2002. PMID: 11943137 Free PMC article.
-
Glycogen synthase kinase-3 regulates endoplasmic reticulum (ER) stress-induced CHOP expression in neuronal cells.Exp Cell Res. 2011 Jul 1;317(11):1621-8. doi: 10.1016/j.yexcr.2011.02.012. Epub 2011 Feb 26. Exp Cell Res. 2011. PMID: 21356208 Free PMC article.
-
The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends.J Biol Chem. 2019 Feb 8;294(6):2098-2108. doi: 10.1074/jbc.REV118.002804. Epub 2018 Dec 18. J Biol Chem. 2019. PMID: 30563838 Free PMC article. Review.
-
GRP78 in lung cancer.J Transl Med. 2021 Mar 21;19(1):118. doi: 10.1186/s12967-021-02786-6. J Transl Med. 2021. PMID: 33743739 Free PMC article. Review.
Cited by
-
ER stress-mediated apoptosis induced by celastrol in cancer cells and important role of glycogen synthase kinase-3β in the signal network.Cell Death Dis. 2013 Jul 11;4(7):e715. doi: 10.1038/cddis.2013.222. Cell Death Dis. 2013. PMID: 23846217 Free PMC article.
-
Muscle wasting and the temporal gene expression pattern in a novel rat intensive care unit model.BMC Genomics. 2011 Dec 13;12:602. doi: 10.1186/1471-2164-12-602. BMC Genomics. 2011. PMID: 22165895 Free PMC article.
-
Carfilzomib Promotes the Unfolded Protein Response and Apoptosis in Cetuximab-Resistant Colorectal Cancer.Int J Mol Sci. 2021 Jul 1;22(13):7114. doi: 10.3390/ijms22137114. Int J Mol Sci. 2021. PMID: 34281166 Free PMC article.
-
Identification of O-GlcNAc modification targets in mouse retinal pericytes: implication of p53 in pathogenesis of diabetic retinopathy.PLoS One. 2014 May 1;9(5):e95561. doi: 10.1371/journal.pone.0095561. eCollection 2014. PLoS One. 2014. PMID: 24788674 Free PMC article.
-
Hepatitis B Virus DNA Polymerase Displays an Anti-Apoptotic Effect by Interacting with Elongation Factor-1 Alpha-2 in Hepatoma Cells.J Microbiol Biotechnol. 2021 Jan 28;31(1):16-24. doi: 10.4014/jmb.2002.02039. J Microbiol Biotechnol. 2021. PMID: 33144545 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

