Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor
- PMID: 18073109
- PMCID: PMC2217667
- DOI: 10.1016/j.str.2007.10.014
Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor
Abstract
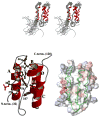
Phosphorylation of endogenous inhibitor proteins for type-1 Ser/Thr phosphatase (PP1) provides a mechanism for reciprocal coordination of kinase and phosphatase activities. A myosin phosphatase inhibitor protein CPI-17 is phosphorylated at Thr38 through G-protein-mediated signals, resulting in a >1000-fold increase in inhibitory potency. We show here the solution NMR structure of phospho-T38-CPI-17 with rmsd of 0.36 +/- 0.06 A for the backbone secondary structure, which reveals how phosphorylation triggers a conformational change and exposes an inhibitory surface. This active conformation is stabilized by the formation of a hydrophobic core of intercalated side chains, which is not formed in a phospho-mimetic D38 form of CPI-17. Thus, the profound increase in potency of CPI-17 arises from phosphorylation, conformational change, and hydrophobic stabilization of a rigid structure that poses the phosphorylated residue on the protein surface and restricts its hydrolysis by myosin phosphatase. Our results provide structural insights into transduction of kinase signals by PP1 inhibitor proteins.
Figures


Similar articles
-
Solution NMR structure of the myosin phosphatase inhibitor protein CPI-17 shows phosphorylation-induced conformational changes responsible for activation.J Mol Biol. 2001 Dec 7;314(4):839-49. doi: 10.1006/jmbi.2001.5200. J Mol Biol. 2001. PMID: 11734001
-
Distinctive solution conformation of phosphatase inhibitor CPI-17 substituted with aspartate at the phosphorylation-site threonine residue.J Mol Biol. 2003 Mar 7;326(5):1539-47. doi: 10.1016/s0022-2836(03)00048-2. J Mol Biol. 2003. PMID: 12595264
-
Phosphoprotein inhibitor CPI-17 specificity depends on allosteric regulation of protein phosphatase-1 by regulatory subunits.Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8888-93. doi: 10.1073/pnas.0307812101. Epub 2004 Jun 7. Proc Natl Acad Sci U S A. 2004. PMID: 15184667 Free PMC article.
-
Histamine-induced vasoconstriction involves phosphorylation of a specific inhibitor protein for myosin phosphatase by protein kinase C alpha and delta isoforms.J Biol Chem. 2001 Aug 3;276(31):29072-8. doi: 10.1074/jbc.M103206200. Epub 2001 Jun 7. J Biol Chem. 2001. PMID: 11397799
-
Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors.J Biol Chem. 2009 Dec 18;284(51):35273-7. doi: 10.1074/jbc.R109.059972. J Biol Chem. 2009. PMID: 19846560 Free PMC article. Review.
Cited by
-
Possible roles of N- and C-terminal unstructured tails of CPI-17 in regulating Ca2+ sensitization force of smooth muscle.J Smooth Muscle Res. 2022;58(0):22-33. doi: 10.1540/jsmr.58.22. J Smooth Muscle Res. 2022. PMID: 35418530 Free PMC article.
-
The actin cytoskeleton in cancer cell motility.Clin Exp Metastasis. 2009;26(4):273-87. doi: 10.1007/s10585-008-9174-2. Epub 2008 May 23. Clin Exp Metastasis. 2009. PMID: 18498004 Review.
-
Sevoflurane and isoflurane inhibit KCl-induced Class II phosphoinositide 3-kinase α subunit mediated vasoconstriction in rat aorta.BMC Anesthesiol. 2016 Aug 18;16(1):63. doi: 10.1186/s12871-016-0227-9. BMC Anesthesiol. 2016. PMID: 27538808 Free PMC article.
-
New insights into RhoA/Rho-kinase signaling: a key regulator of vascular contraction.Small GTPases. 2021 Sep-Nov;12(5-6):458-469. doi: 10.1080/21541248.2020.1822721. Epub 2020 Sep 24. Small GTPases. 2021. PMID: 32970516 Free PMC article. Review.
-
Solution structure of the inhibitory phosphorylation domain of myosin phosphatase targeting subunit 1.Proteins. 2009 Nov 15;77(3):732-5. doi: 10.1002/prot.22529. Proteins. 2009. PMID: 19701943 Free PMC article. No abstract available.
References
-
- Bax A, Pochapsky SS. Optimized recording of heteronuclear multidimensional NMR spectra using pulsed field gradients. J Magn Reson. 1992;99:638–643.
-
- Bonnevier J, Arner A. Actions downstream of cyclic GMP/protein kinase G can reverse protein kinase C-mediated phosphorylation of CPI-17 and Ca(2+) sensitization in smooth muscle. J Biol Chem. 2004;279:28998–29003. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat Struct Mol Biol. 2005;12:987–993. - PubMed
-
- Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

