Evidence against roles for phorbol binding protein Munc13-1, ADAM adaptor Eve-1, or vesicle trafficking phosphoproteins Munc18 or NSF as phospho-state-sensitive modulators of phorbol/PKC-activated Alzheimer APP ectodomain shedding
- PMID: 18067682
- PMCID: PMC2211485
- DOI: 10.1186/1750-1326-2-23
Evidence against roles for phorbol binding protein Munc13-1, ADAM adaptor Eve-1, or vesicle trafficking phosphoproteins Munc18 or NSF as phospho-state-sensitive modulators of phorbol/PKC-activated Alzheimer APP ectodomain shedding
Abstract
Background: Shedding of the Alzheimer amyloid precursor protein (APP) ectodomain can be accelerated by phorbol esters, compounds that act via protein kinase C (PKC) or through unconventional phorbol-binding proteins such as Munc13-1. We have previously demonstrated that application of phorbol esters or purified PKC potentiates budding of APP-bearing secretory vesicles at the trans-Golgi network (TGN) and toward the plasma membrane where APP becomes a substrate for enzymes responsible for shedding, known collectively as alpha-secretase(s). However, molecular identification of the presumptive "phospho-state-sensitive modulators of ectodomain shedding" (PMES) responsible for regulated shedding has been challenging. Here, we examined the effects on APP ectodomain shedding of four phorbol-sensitive proteins involved in regulation of vesicular membrane trafficking of APP: Munc13-1, Munc18, NSF, and Eve-1.
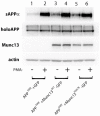
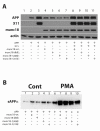
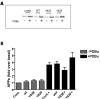
Results: Overexpression of either phorbol-sensitive wildtype Munc13-1 or phorbol-insensitive Munc13-1 H567K resulted in increased basal APP ectodomain shedding. However, in contrast to the report of Rossner et al (2004), phorbol ester-dependent APP ectodomain shedding from cells overexpressing APP and Munc13-1 wildtype was indistinguishable from that observed following application of phorbol to cells overexpressing APP and Munc13-1 H567K mutant. This pattern of similar effects on basal and stimulated APP shedding was also observed for Munc18 and NSF. Eve-1, an ADAM adaptor protein reported to be essential for PKC-regulated shedding of pro-EGF, was found to play no obvious role in regulated shedding of sAPPalpha.
Conclusion: Our results indicate that, in the HEK293 system, Munc13-1, Munc18, NSF, and EVE-1 fail to meet essential criteria for identity as PMES for APP.
Figures

Similar articles
-
Phorbol esters modulate spontaneous and Ca2+-evoked transmitter release via acting on both Munc13 and protein kinase C.J Neurosci. 2008 Aug 13;28(33):8257-67. doi: 10.1523/JNEUROSCI.0550-08.2008. J Neurosci. 2008. PMID: 18701688 Free PMC article.
-
Atorvastatin-induced activation of Alzheimer's alpha secretase is resistant to standard inhibitors of protein phosphorylation-regulated ectodomain shedding.J Neurochem. 2004 Aug;90(4):1005-10. doi: 10.1111/j.1471-4159.2004.02521.x. J Neurochem. 2004. PMID: 15287907
-
Inhibition of dynamin-dependent endocytosis increases shedding of the amyloid precursor protein ectodomain and reduces generation of amyloid beta protein.BMC Cell Biol. 2005 Aug 11;6:30. doi: 10.1186/1471-2121-6-30. BMC Cell Biol. 2005. PMID: 16095541 Free PMC article.
-
Regulated cleavage of the Alzheimer amyloid precursor protein: molecular and cellular basis.Biochimie. 1994;76(3-4):300-3. doi: 10.1016/0300-9084(94)90162-7. Biochimie. 1994. PMID: 7819339 Review.
-
New players in old amyloid precursor protein-processing pathways.Int J Dev Neurosci. 2004 Nov;22(7):467-74. doi: 10.1016/j.ijdevneu.2004.07.004. Int J Dev Neurosci. 2004. PMID: 15465276 Review.
Cited by
-
Bryostatin-1 vs. TPPB: dose-dependent APP processing and PKC-α, -δ, and -ε isoform activation in SH-SY5Y neuronal cells.J Mol Neurosci. 2012 Sep;48(1):234-44. doi: 10.1007/s12031-012-9816-3. Epub 2012 Jun 15. J Mol Neurosci. 2012. PMID: 22700373 Free PMC article.
-
Munc13 Is a Molecular Target of Bryostatin 1.Biochemistry. 2019 Jul 9;58(27):3016-3030. doi: 10.1021/acs.biochem.9b00427. Epub 2019 Jun 20. Biochemistry. 2019. PMID: 31243993 Free PMC article.
-
Resveratrol inhibits phorbol ester-induced membrane translocation of presynaptic Munc13-1.Biochim Biophys Acta Gen Subj. 2017 Nov;1861(11 Pt A):2640-2651. doi: 10.1016/j.bbagen.2017.07.006. Epub 2017 Jul 13. Biochim Biophys Acta Gen Subj. 2017. PMID: 28713022 Free PMC article.
-
Protein kinase C and rho activated coiled coil protein kinase 2 (ROCK2) modulate Alzheimer's APP metabolism and phosphorylation of the Vps10-domain protein, SorL1.Mol Neurodegener. 2010 Dec 30;5:62. doi: 10.1186/1750-1326-5-62. Mol Neurodegener. 2010. PMID: 21192821 Free PMC article.
-
Enhanced generation of Alzheimer's amyloid-beta following chronic exposure to phorbol ester correlates with differential effects on alpha and epsilon isozymes of protein kinase C.J Neurochem. 2009 Jan;108(2):319-30. doi: 10.1111/j.1471-4159.2008.05770.x. Epub 2008 Dec 2. J Neurochem. 2009. PMID: 19012746 Free PMC article.
References
-
- Buxbaum JD, Gandy SE, Cicchetti P, Ehrlich ME, Czernik AJ, Fracasso RP, Ramabhadran TV, Unterbeck AJ, Greengard P. Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. - DOI - PMC - PubMed
-
- Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, Johnson RS, Castner BJ, Cerretti DP, Black RA. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. - DOI - PubMed
-
- Koike H, Tomioka S, Sorimachi H, Saido TC, Maruyama K, Okuyama A, Fujisawa-Sehara A, Ohno S, Suzuki K, Ishiura S. Membrane-anchored metalloprotease MDC9 has an alpha-secretase activity responsible for processing the amyloid precursor protein. Biochem J. 1999;343:371–375. doi: 10.1042/0264-6021:3430371. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

