Insights into kinetochore-DNA interactions from the structure of Cep3Delta
- PMID: 18064045
- PMCID: PMC2246632
- DOI: 10.1038/sj.embor.7401139
Insights into kinetochore-DNA interactions from the structure of Cep3Delta
Abstract
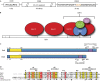
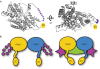
The CBF3 complex is an essential core component of the budding yeast kinetochore and is required for the centromeric localization of all other kinetochore proteins. We determined the crystal structure of a large section of the protein Cep3 from CBF3, which is the only component with obvious DNA-binding motifs. The protein adopts a roughly bilobal shape, with an extended dimerization interface. The dimer has a large central channel that is sufficient to accommodate duplex B-form DNA. The zinc-finger domains emerge at the edges of the channel, and could bind to the DNA in a pseudo-symmetrical manner at degenerate half-sites in the centromeric sequence. We propose a mechanism for the modulation of DNA affinity by an acidic activator domain, which could be applicable to a wider family of transcription factors.
Figures
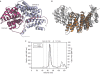


Similar articles
-
Crystal structure of the yeast inner kinetochore subunit Cep3p.Structure. 2007 Nov;15(11):1422-30. doi: 10.1016/j.str.2007.09.008. Structure. 2007. PMID: 17997968 Free PMC article.
-
The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex.J Cell Biol. 1999 May 31;145(5):933-50. doi: 10.1083/jcb.145.5.933. J Cell Biol. 1999. PMID: 10352012 Free PMC article.
-
Architecture of the CBF3-centromere complex of the budding yeast kinetochore.Nat Struct Mol Biol. 2018 Dec;25(12):1103-1110. doi: 10.1038/s41594-018-0154-1. Epub 2018 Nov 26. Nat Struct Mol Biol. 2018. PMID: 30478265 Free PMC article.
-
The Saccharomyces cerevisiae kinetochore.FEBS Lett. 1996 Jun 24;389(1):70-4. doi: 10.1016/0014-5793(96)00563-7. FEBS Lett. 1996. PMID: 8682209 Review.
-
Structures and functions of yeast kinetochore complexes.Annu Rev Biochem. 2007;76:563-91. doi: 10.1146/annurev.biochem.76.052705.160607. Annu Rev Biochem. 2007. PMID: 17362199 Review.
Cited by
-
DNA damage signalling targets the kinetochore to promote chromatin mobility.Nat Cell Biol. 2016 Mar;18(3):281-90. doi: 10.1038/ncb3308. Epub 2016 Feb 1. Nat Cell Biol. 2016. PMID: 26829389
-
Structural basis for assembly of the CBF3 kinetochore complex.EMBO J. 2018 Jan 17;37(2):269-281. doi: 10.15252/embj.201798134. Epub 2017 Dec 6. EMBO J. 2018. PMID: 29212814 Free PMC article.
-
The Role of the Fusarium oxysporum FTF2 Transcription Factor in Host Colonization and Virulence in Common Bean Plants (Phaseolus vulgaris L.).Pathogens. 2023 Feb 26;12(3):380. doi: 10.3390/pathogens12030380. Pathogens. 2023. PMID: 36986302 Free PMC article.
-
CeGAL: Redefining a Widespread Fungal-Specific Transcription Factor Family Using an In Silico Error-Tracking Approach.J Fungi (Basel). 2023 Mar 29;9(4):424. doi: 10.3390/jof9040424. J Fungi (Basel). 2023. PMID: 37108879 Free PMC article.
-
The composition, functions, and regulation of the budding yeast kinetochore.Genetics. 2013 Aug;194(4):817-46. doi: 10.1534/genetics.112.145276. Genetics. 2013. PMID: 23908374 Free PMC article. Review.
References
-
- Andrade M, Bork P (1995) HEAT repeats in the Huntington's disease protein. Nat Genet 11: 115–116 - PubMed
-
- Carbon J, Clarke L (1984) Structural and functional analysis of a yeast centromere (CEN3). J Cell Sci Suppl 1: 43–58 - PubMed
-
- Clarke L, Carbon J (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

