Identification of novel modulators of mitochondrial function by a genome-wide RNAi screen in Drosophila melanogaster
- PMID: 18042644
- PMCID: PMC2134776
- DOI: 10.1101/gr.6940108
Identification of novel modulators of mitochondrial function by a genome-wide RNAi screen in Drosophila melanogaster
Abstract
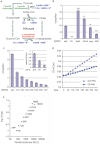
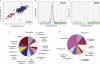
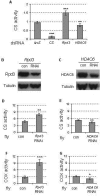
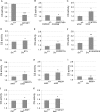
Mitochondrial dysfunction is associated with many human diseases. There has not been a systematic genetic approach for identifying regulators of basal mitochondrial biogenesis and function in higher eukaryotes. We performed a genome-wide RNA interference (RNAi) screen in Drosophila cells using mitochondrial Citrate synthase (CS) activity as the primary readout. We screened 13,071 dsRNAs and identified 152 genes that modulate CS activity. These modulators are involved in a wide range of biological processes and pathways including mitochondrial-related functions, transcriptional and translational regulation, and signaling pathways. Selected hits among the 152 genes were further analyzed for their effect on mitochondrial CS activity in transgenic flies or fly mutants. We confirmed a number of gene hits including HDAC6, Rpd3(HDAC1), CG3249, vimar, Src42A, klumpfuss, barren, and smt3 which exert effects on mitochondrial CS activities in vivo, demonstrating the value of Drosophila genome-wide RNAi screens for identifying genes and pathways that modulate mitochondrial function.
Figures
Similar articles
-
Nuclear genomic control of naturally occurring variation in mitochondrial function in Drosophila melanogaster.BMC Genomics. 2012 Nov 22;13:659. doi: 10.1186/1471-2164-13-659. BMC Genomics. 2012. PMID: 23171078 Free PMC article.
-
Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi.Nature. 2009 Apr 23;458(7241):987-92. doi: 10.1038/nature07936. Epub 2009 Apr 12. Nature. 2009. PMID: 19363474 Free PMC article.
-
Advances in genome-wide RNAi cellular screens: a case study using the Drosophila JAK/STAT pathway.BMC Genomics. 2012 Sep 24;13:506. doi: 10.1186/1471-2164-13-506. BMC Genomics. 2012. PMID: 23006893 Free PMC article.
-
[Tissue-specific genome-wide RNAi screen in Drosophila].Tanpakushitsu Kakusan Koso. 2010 Jan;55(1):26-33. Tanpakushitsu Kakusan Koso. 2010. PMID: 20058703 Review. Japanese. No abstract available.
-
A Guide to Genome-Wide In Vivo RNAi Applications in Drosophila.Methods Mol Biol. 2016;1478:117-143. doi: 10.1007/978-1-4939-6371-3_6. Methods Mol Biol. 2016. PMID: 27730578 Review.
Cited by
-
cAMP and mitochondria.Physiology (Bethesda). 2013 May;28(3):199-209. doi: 10.1152/physiol.00004.2013. Physiology (Bethesda). 2013. PMID: 23636265 Free PMC article. Review.
-
Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice.Free Radic Biol Med. 2012 Oct 15;53(8):1584-94. doi: 10.1016/j.freeradbiomed.2012.08.009. Epub 2012 Aug 23. Free Radic Biol Med. 2012. PMID: 22940620 Free PMC article.
-
Rapid and transient oxygen consumption increase following acute HDAC/KDAC inhibition in Drosophila tissue.Sci Rep. 2018 Mar 8;8(1):4199. doi: 10.1038/s41598-018-22674-2. Sci Rep. 2018. PMID: 29520020 Free PMC article.
-
Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile.BMC Cell Biol. 2012 Oct 29;13:26. doi: 10.1186/1471-2121-13-26. BMC Cell Biol. 2012. PMID: 23107381 Free PMC article.
-
The mitochondrial chaperone protein TRAP1 mitigates α-Synuclein toxicity.PLoS Genet. 2012 Feb;8(2):e1002488. doi: 10.1371/journal.pgen.1002488. Epub 2012 Feb 2. PLoS Genet. 2012. PMID: 22319455 Free PMC article.
References
-
- Affaitati A., Cardone L., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Cardone L., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., De C.T., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Carlucci A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Ginsberg M.D., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Varrone S., Gottesman M.E., Avvedimento E.V., Feliciello A., Gottesman M.E., Avvedimento E.V., Feliciello A., Avvedimento E.V., Feliciello A., Feliciello A. Essential role of A-kinase anchor protein 121 for cAMP signaling to mitochondria. J. Biol. Chem. 2003;278:4286–4294. - PubMed
-
- Appaix F., Kuznetsov A.V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kuznetsov A.V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Usson Y., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kay L., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Andrienko T., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Olivares J., Kaambre T., Sikk P., Margreiter R., Saks V., Kaambre T., Sikk P., Margreiter R., Saks V., Sikk P., Margreiter R., Saks V., Margreiter R., Saks V., Saks V. Possible role of cytoskeleton in intracellular arrangement and regulation of mitochondria. Exp. Physiol. 2003;88:175–190. - PubMed
-
- Bakin R.E., Jung M.O., Jung M.O. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J. Biol. Chem. 2004;279:51218–51225. - PubMed
-
- Barlow A.L., van Drunen C.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., van Drunen C.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., Johnson C.A., Tweedie S., Bird A., Turner B.M., Tweedie S., Bird A., Turner B.M., Bird A., Turner B.M., Turner B.M. dSIR2 and dHDAC6: Two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 2001;265:90–103. - PubMed
-
- Bhat M.A., Philp A.V., Glover D.M., Bellen H.J., Philp A.V., Glover D.M., Bellen H.J., Glover D.M., Bellen H.J., Bellen H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
