Adenovirus RIDalpha regulates endosome maturation by mimicking GTP-Rab7
- PMID: 18039930
- PMCID: PMC2099200
- DOI: 10.1083/jcb.200702187
Adenovirus RIDalpha regulates endosome maturation by mimicking GTP-Rab7
Abstract
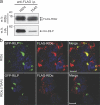
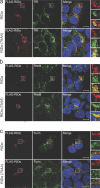
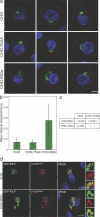
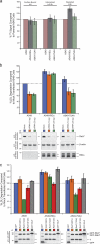
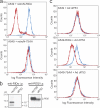
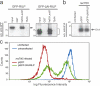
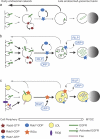
The small guanosine triphosphatase Rab7 regulates late endocytic trafficking. Rab7-interacting lysosomal protein (RILP) and oxysterol-binding protein-related protein 1L (ORP1L) are guanosine triphosphate (GTP)-Rab7 effectors that instigate minus end-directed microtubule transport. We demonstrate that RILP and ORP1L both interact with the group C adenovirus protein known as receptor internalization and degradation alpha (RIDalpha), which was previously shown to clear the cell surface of several membrane proteins, including the epidermal growth factor receptor and Fas (Carlin, C.R., A.E. Tollefson, H.A. Brady, B.L. Hoffman, and W.S. Wold. 1989. Cell. 57:135-144; Shisler, J., C. Yang, B. Walter, C.F. Ware, and L.R. Gooding. 1997. J. Virol. 71:8299-8306). RIDalpha localizes to endocytic vesicles but is not homologous to Rab7 and is not catalytically active. We show that RIDalpha compensates for reduced Rab7 or dominant-negative (DN) Rab7(T22N) expression. In vitro, Cu(2+) binding to RIDalpha residues His75 and His76 facilitates the RILP interaction. Site-directed mutagenesis of these His residues results in the loss of RIDalpha-RILP interaction and RIDalpha activity in cells. Additionally, expression of the RILP DN C-terminal region hinders RIDalpha activity during an acute adenovirus infection. We conclude that RIDalpha coordinates recruitment of these GTP-Rab7 effectors to compartments that would ordinarily be perceived as early endosomes, thereby promoting the degradation of selected cargo.
Figures



Similar articles
-
Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin.J Cell Biol. 2007 Feb 12;176(4):459-71. doi: 10.1083/jcb.200606077. Epub 2007 Feb 5. J Cell Biol. 2007. PMID: 17283181 Free PMC article.
-
A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning.J Biol Chem. 2018 Sep 7;293(36):14155-14164. doi: 10.1074/jbc.RA118.001854. Epub 2018 Jul 16. J Biol Chem. 2018. PMID: 30012887 Free PMC article.
-
Rab24 interacts with the Rab7/Rab interacting lysosomal protein complex to regulate endosomal degradation.Traffic. 2016 Nov;17(11):1181-1196. doi: 10.1111/tra.12431. Epub 2016 Oct 3. Traffic. 2016. PMID: 27550070
-
Rab7: role of its protein interaction cascades in endo-lysosomal traffic.Cell Signal. 2011 Mar;23(3):516-21. doi: 10.1016/j.cellsig.2010.09.012. Epub 2010 Sep 21. Cell Signal. 2011. PMID: 20851765 Review.
-
Regulation of Endosomal Trafficking by Rab7 and Its Effectors in Neurons: Clues from Charcot-Marie-Tooth 2B Disease.Biomolecules. 2023 Sep 16;13(9):1399. doi: 10.3390/biom13091399. Biomolecules. 2023. PMID: 37759799 Free PMC article. Review.
Cited by
-
Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration.J Biol Chem. 2009 May 1;284(18):12110-24. doi: 10.1074/jbc.M809277200. Epub 2009 Mar 5. J Biol Chem. 2009. PMID: 19265192 Free PMC article.
-
New Insights to Adenovirus-Directed Innate Immunity in Respiratory Epithelial Cells.Microorganisms. 2019 Jul 25;7(8):216. doi: 10.3390/microorganisms7080216. Microorganisms. 2019. PMID: 31349602 Free PMC article. Review.
-
Adenovirus RID-alpha activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C.J Cell Biol. 2009 Nov 16;187(4):537-52. doi: 10.1083/jcb.200903039. J Cell Biol. 2009. PMID: 19948501 Free PMC article.
-
Adenovirus early region 3 RIDα protein limits NFκB signaling through stress-activated EGF receptors.PLoS Pathog. 2019 Aug 19;15(8):e1008017. doi: 10.1371/journal.ppat.1008017. eCollection 2019 Aug. PLoS Pathog. 2019. PMID: 31425554 Free PMC article.
-
Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain.Mol Biol Cell. 2010 Dec;21(23):4162-72. doi: 10.1091/mbc.E10-06-0495. Epub 2010 Oct 13. Mol Biol Cell. 2010. PMID: 20943950 Free PMC article.
References
-
- Agarraberes, F.A., and J.F. Dice. 2001. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114:2491–2499. - PubMed
-
- Brown, M.S., and J.L. Goldstein. 1975. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell. 6:307–316. - PubMed
-
- Bucci, C., R.G. Parton, I.H. Mather, H. Stunnenberg, K. Simons, B. Hoflack, and M. Zerial. 1992. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 70:715–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

