Uncoupling RNA virus replication from transcription via the polymerase: functional and evolutionary insights
- PMID: 18034156
- PMCID: PMC2140117
- DOI: 10.1038/sj.emboj.7601931
Uncoupling RNA virus replication from transcription via the polymerase: functional and evolutionary insights
Abstract
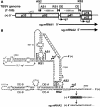
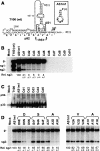
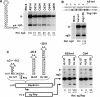
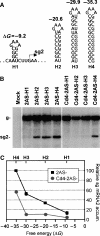
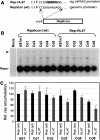
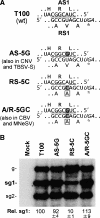
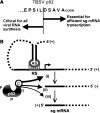
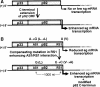
Many eukaryotic positive-strand RNA viruses transcribe subgenomic (sg) mRNAs that are virus-derived messages that template the translation of a subset of viral proteins. Currently, the premature termination (PT) mechanism of sg mRNA transcription, a process thought to operate in a variety of viruses, is best understood in tombusviruses. The viral RNA elements involved in regulating this mechanism have been well characterized in several systems; however, no corresponding protein factors have been identified yet. Here we show that tombusvirus genome replication can be effectively uncoupled from sg mRNA transcription in vivo by C-terminal modifications in its RNA-dependent RNA polymerase (RdRp). Systematic analysis of the PT transcriptional pathway using viral genomes harboring mutant RdRps revealed that the C-terminus functions primarily at an early step in this mechanism by mediating both efficient and accurate production of minus-strand templates for sg mRNA transcription. Our results also suggest a simple evolutionary scheme by which the virus could gain or enhance its transcriptional activity, and define global folding of the viral RNA genome as a previously unappreciated determinant of RdRp evolution.
Figures


Similar articles
-
Conserved motifs in a tombusvirus polymerase modulate genome replication, subgenomic transcription, and amplification of defective interfering RNAs.J Virol. 2015 Mar;89(6):3236-46. doi: 10.1128/JVI.03378-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568204 Free PMC article.
-
A multicomponent RNA-based control system regulates subgenomic mRNA transcription in a tombusvirus.J Virol. 2007 Mar;81(5):2429-39. doi: 10.1128/JVI.01969-06. Epub 2006 Dec 13. J Virol. 2007. PMID: 17166897 Free PMC article.
-
Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro.J Virol. 2012 Jan;86(1):156-71. doi: 10.1128/JVI.00404-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013057 Free PMC article.
-
Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle.Virology. 2011 Apr 10;412(2):245-55. doi: 10.1016/j.virol.2011.02.007. Epub 2011 Mar 5. Virology. 2011. PMID: 21377709 Free PMC article. Review.
-
Tombusvirus polymerase: Structure and function.Virus Res. 2017 Apr 15;234:74-86. doi: 10.1016/j.virusres.2017.01.012. Epub 2017 Jan 19. Virus Res. 2017. PMID: 28111194 Review.
Cited by
-
Dimerization of an umbravirus RNA genome activates subgenomic mRNA transcription.Nucleic Acids Res. 2023 Sep 8;51(16):8787-8804. doi: 10.1093/nar/gkad550. Nucleic Acids Res. 2023. PMID: 37395397 Free PMC article.
-
Higher-order RNA structural requirements and small-molecule induction of tombusvirus subgenomic mRNA transcription.J Virol. 2008 Apr;82(8):3864-71. doi: 10.1128/JVI.02416-07. Epub 2008 Feb 6. J Virol. 2008. PMID: 18256151 Free PMC article.
-
Analytical validation of quantitative SARS-CoV-2 subgenomic and viral load laboratory developed tests conducted on the Panther Fusion® (Hologic) with preliminary application to clinical samples.PLoS One. 2023 Jun 29;18(6):e0287576. doi: 10.1371/journal.pone.0287576. eCollection 2023. PLoS One. 2023. PMID: 37384714 Free PMC article.
-
Conserved motifs in a tombusvirus polymerase modulate genome replication, subgenomic transcription, and amplification of defective interfering RNAs.J Virol. 2015 Mar;89(6):3236-46. doi: 10.1128/JVI.03378-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568204 Free PMC article.
-
A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes.PLoS Pathog. 2009 Mar;5(3):e1000323. doi: 10.1371/journal.ppat.1000323. Epub 2009 Mar 6. PLoS Pathog. 2009. PMID: 19266082 Free PMC article.
References
-
- Choi IR, Ostrovsky M, Zhang G, White KA (2001) Regulatory activity of distal and core RNA elements in Tombusvirus subgenomic mRNA2 transcription. J Biol Chem 276: 41761–41768 - PubMed
-
- Choi IR, White KA (2002) An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J Biol Chem 277: 3760–3766 - PubMed
-
- Cowley JA, Dimmock CM, Walker PJ (2002) Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J Gen Virol 83: 927–935 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

