The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation
- PMID: 18025255
- PMCID: PMC2151041
- DOI: 10.1261/rna.777308
The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation
Abstract
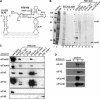
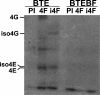
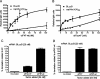
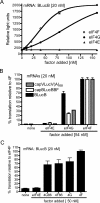
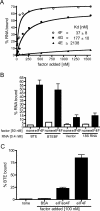
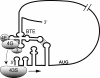
The 3' cap-independent translation element (BTE) of Barley yellow dwarf virus RNA confers efficient translation initiation at the 5' end via long-distance base pairing with the 5'-untranslated region (UTR). Here we provide evidence that the BTE functions by recruiting translation initiation factor eIF4F. We show that the BTE interacts specifically with the cap-binding initiation factor complexes eIF4F and eIFiso4F in a wheat germ extract (wge). In wge depleted of cap-interacting factors, addition of eIF4F (and to a lesser extent, eIFiso4F) allowed efficient translation of an uncapped reporter construct (BLucB) containing the BTE in its 3' UTR. Translation of BLucB required much lower levels of eIF4F or eIFiso4F than did a capped, nonviral mRNA. Both full-length eIF4G and the carboxy-terminal half of eIF4G lacking the eIF4E binding site stimulated translation to 70% of the level obtained with eIF4F, indicating a minor role for the cap-binding protein, eIF4E. In wge inhibited by either BTE in trans or cap analog, eIF4G alone restored translation nearly as much as eIF4F, while addition of eIF4E alone had no effect. The BTE bound eIF4G (Kd = 177 nm) and eIF4F (Kd = 37 nm) with high affinity, but very weakly to eIF4E. These interactions correlate with the ability of the factors to facilitate BTE-mediated translation. These results and previous observations are consistent with a model in which eIF4F is delivered to the 5' UTR by the BTE, and they show that eIF4G, but not eIF4E, plays a major role in this novel mechanism of cap-independent translation.
Figures

Similar articles
-
Recruitment of the 40S ribosome subunit to the 3'-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation.J Biol Chem. 2015 May 1;290(18):11268-81. doi: 10.1074/jbc.M115.645002. Epub 2015 Mar 19. J Biol Chem. 2015. PMID: 25792742 Free PMC article.
-
Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3' cap-independent translation element (BTE).J Biol Chem. 2017 Apr 7;292(14):5921-5931. doi: 10.1074/jbc.M116.764902. Epub 2017 Feb 27. J Biol Chem. 2017. PMID: 28242763 Free PMC article.
-
Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3'-untranslated region correlates with translation efficiency.J Biol Chem. 2014 Feb 14;289(7):4286-94. doi: 10.1074/jbc.M113.530329. Epub 2013 Dec 30. J Biol Chem. 2014. PMID: 24379412 Free PMC article.
-
Manipulation of the host translation initiation complex eIF4F by DNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1511-6. doi: 10.1042/BST0381511. Biochem Soc Trans. 2010. PMID: 21118117 Review.
-
eIF4F: a retrospective.J Biol Chem. 2015 Oct 2;290(40):24091-9. doi: 10.1074/jbc.R115.675280. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324716 Free PMC article. Review.
Cited by
-
Eukaryotic initiation factor (eIF) 3 mediates Barley Yellow Dwarf Viral mRNA 3'-5' UTR interactions and 40S ribosomal subunit binding to facilitate cap-independent translation.Nucleic Acids Res. 2019 Jul 9;47(12):6225-6235. doi: 10.1093/nar/gkz448. Nucleic Acids Res. 2019. PMID: 31114905 Free PMC article.
-
A Dual Interaction Between the 5'- and 3'-Ends of the Melon Necrotic Spot Virus (MNSV) RNA Genome Is Required for Efficient Cap-Independent Translation.Front Plant Sci. 2018 May 9;9:625. doi: 10.3389/fpls.2018.00625. eCollection 2018. Front Plant Sci. 2018. PMID: 29868081 Free PMC article.
-
The RNA of Maize Chlorotic Mottle Virus, an Obligatory Component of Maize Lethal Necrosis Disease, Is Translated via a Variant Panicum Mosaic Virus-Like Cap-Independent Translation Element.J Virol. 2020 Oct 27;94(22):e01005-20. doi: 10.1128/JVI.01005-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847851 Free PMC article.
-
Recruitment of the 40S ribosome subunit to the 3'-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation.J Biol Chem. 2015 May 1;290(18):11268-81. doi: 10.1074/jbc.M115.645002. Epub 2015 Mar 19. J Biol Chem. 2015. PMID: 25792742 Free PMC article.
-
Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus.Virology. 2008 Jun 5;375(2):354-60. doi: 10.1016/j.virol.2008.01.035. Epub 2008 Mar 7. Virology. 2008. PMID: 18329064 Free PMC article.
References
-
- Allen, M.L., Metz, A.M., Timmer, R.T., Rhoads, R.E., Browning, K.S. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J. Biol. Chem. 1992;267:23232–23236. - PubMed
-
- Allen, E., Wang, S., Miller, W.A. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology. 1999;253:139–144. - PubMed
-
- Basso, J., Dallaire, P., Charest, P.J., Devantier, Y., Laliberte, J.F. Evidence for an internal ribosome entry site within the 5′ nontranslated region of turnip mosaic potyvirus RNA. J. Gen. Virol. 1994;75:3157–3165. - PubMed
-
- Batten, J.S., Desvoyes, B., Yamamura, Y., Scholthof, K.B. A translational enhancer element on the 3′-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580:2591–2597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
