Transcriptional homogenization of rDNA repeats in the episome-based nucleolus induces genome-wide changes in the chromosomal distribution of condensin
- PMID: 18023874
- PMCID: PMC2366798
- DOI: 10.1016/j.plasmid.2007.09.003
Transcriptional homogenization of rDNA repeats in the episome-based nucleolus induces genome-wide changes in the chromosomal distribution of condensin
Abstract
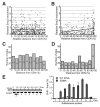
Condensin activity establishes and maintains mitotic chromosome condensation, however the mechanisms of condensin recognition of specific chromosomal sites remain unknown. rDNA is the chief condensin binding locus in Saccharomyces cerevisiae, and the level of nucleolar transcription is one of the key factors determining condensin loading to the nucleolar organizer. A new aspect of this transcriptional control is demonstrated in cells with a diffuse (episomal) nucleolar organizer, where active transcription excludes condensin from the transcribed regions of rDNA. Genome-wide ChIP-chip analysis showed that these cells acquire an altered and a more robust pattern of chromosomal condensin distribution, with increased enrichment of wild-type hotspots and with emergence of new sites, most notably in the subtelomeric regions. This genome-wide condensin relocalization induced by the increase in rDNA transcription and, possibly, nucleolar architecture uncovers a novel potential role of the nucleolus in the general chromosome organization.
Figures


Similar articles
-
Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase.Cell Cycle. 2004 Jul;3(7):960-7. doi: 10.4161/cc.3.7.1003. Epub 2004 Jul 4. Cell Cycle. 2004. PMID: 15190202 Free PMC article.
-
Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription.Cell Cycle. 2006 Oct;5(19):2260-7. doi: 10.4161/cc.5.19.3292. Epub 2006 Oct 1. Cell Cycle. 2006. PMID: 16969110 Free PMC article.
-
Condensin and Hmo1 Mediate a Starvation-Induced Transcriptional Position Effect within the Ribosomal DNA Array.Cell Rep. 2016 Feb 9;14(5):1010-1017. doi: 10.1016/j.celrep.2016.01.005. Epub 2016 Jan 28. Cell Rep. 2016. PMID: 26832415 Free PMC article.
-
Ribosomal DNA and the nucleolus in the context of genome organization.Chromosome Res. 2019 Mar;27(1-2):109-127. doi: 10.1007/s10577-018-9600-5. Epub 2019 Jan 17. Chromosome Res. 2019. PMID: 30656516 Review.
-
Common Features of the Pericentromere and Nucleolus.Genes (Basel). 2019 Dec 10;10(12):1029. doi: 10.3390/genes10121029. Genes (Basel). 2019. PMID: 31835574 Free PMC article. Review.
Cited by
-
Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex.Genetics. 2008 Oct;180(2):715-26. doi: 10.1534/genetics.108.090142. Epub 2008 Sep 9. Genetics. 2008. PMID: 18780748 Free PMC article.
-
High copy and stable expression of the xylanase XynHB in Saccharomyces cerevisiae by rDNA-mediated integration.Sci Rep. 2017 Aug 18;7(1):8747. doi: 10.1038/s41598-017-08647-x. Sci Rep. 2017. PMID: 28821784 Free PMC article.
-
tRNA gene identity affects nuclear positioning.PLoS One. 2011;6(12):e29267. doi: 10.1371/journal.pone.0029267. Epub 2011 Dec 19. PLoS One. 2011. PMID: 22206006 Free PMC article.
-
Nucleoli: composition, function, and dynamics.Plant Physiol. 2012 Jan;158(1):44-51. doi: 10.1104/pp.111.188052. Epub 2011 Nov 14. Plant Physiol. 2012. PMID: 22082506 Free PMC article. Review. No abstract available.
-
Spatial organization of genes as a component of regulated expression.Chromosoma. 2010 Feb;119(1):13-25. doi: 10.1007/s00412-009-0236-2. Epub 2009 Aug 30. Chromosoma. 2010. PMID: 19727792 Free PMC article. Review.
References
-
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. - PubMed
-
- Brown A, Tuite M, editors. Yeast Gene Analysis. Academic Press; San Diego, London: 1998.
-
- D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

