Control of membrane fusion mechanism by lipid composition: predictions from ensemble molecular dynamics
- PMID: 18020701
- PMCID: PMC2077900
- DOI: 10.1371/journal.pcbi.0030220
Control of membrane fusion mechanism by lipid composition: predictions from ensemble molecular dynamics
Abstract
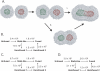
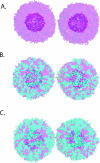
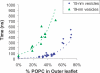
Membrane fusion is critical to biological processes such as viral infection, endocrine hormone secretion, and neurotransmission, yet the precise mechanistic details of the fusion process remain unknown. Current experimental and computational model systems approximate the complex physiological membrane environment for fusion using one or a few protein and lipid species. Here, we report results of a computational model system for fusion in which the ratio of lipid components was systematically varied, using thousands of simulations of up to a microsecond in length to predict the effects of lipid composition on both fusion kinetics and mechanism. In our simulations, increased phosphatidylcholine content in vesicles causes increased activation energies for formation of the initial stalk-like intermediate for fusion and of hemifusion intermediates, in accordance with previous continuum-mechanics theoretical treatments. We also use our large simulation dataset to quantitatively compare the mechanism by which vesicles fuse at different lipid compositions, showing a significant difference in fusion kinetics and mechanism at different compositions simulated. As physiological membranes have different compositions in the inner and outer leaflets, we examine the effect of such asymmetry, as well as the effect of membrane curvature on fusion. These predicted effects of lipid composition on fusion mechanism both underscore the way in which experimental model system construction may affect the observed mechanism of fusion and illustrate a potential mechanism for cellular regulation of the fusion process by altering membrane composition.
Conflict of interest statement
Figures



Similar articles
-
Molecular dynamics simulations of lipid vesicle fusion in atomic detail.Biophys J. 2007 Jun 15;92(12):4254-61. doi: 10.1529/biophysj.106.103572. Epub 2007 Mar 23. Biophys J. 2007. PMID: 17384060 Free PMC article.
-
Initiation and dynamics of hemifusion in lipid bilayers.Biophys J. 2003 Jul;85(1):381-9. doi: 10.1016/S0006-3495(03)74482-8. Biophys J. 2003. PMID: 12829492 Free PMC article.
-
Structure and energy of fusion stalks: the role of membrane edges.Biophys J. 2002 Dec;83(6):2969-80. doi: 10.1016/S0006-3495(02)75303-4. Biophys J. 2002. PMID: 12496070 Free PMC article.
-
The mechanisms of lipid-protein rearrangements during viral infection.Bioelectrochemistry. 2004 Jun;63(1-2):129-36. doi: 10.1016/j.bioelechem.2003.10.016. Bioelectrochemistry. 2004. PMID: 15110263 Review.
-
Membrane fusion: the process and its energy suppliers.Cell Mol Life Sci. 2002 Sep;59(9):1478-90. doi: 10.1007/s00018-002-8523-6. Cell Mol Life Sci. 2002. PMID: 12440770 Free PMC article. Review.
Cited by
-
Function suggests nano-structure: electrophysiology supports that granule membranes play dice.J R Soc Interface. 2012 Oct 7;9(75):2516-26. doi: 10.1098/rsif.2012.0161. Epub 2012 May 23. J R Soc Interface. 2012. PMID: 22628211 Free PMC article.
-
Direct simulation of protein-mediated vesicle fusion: lung surfactant protein B.Biophys J. 2010 Oct 6;99(7):2134-42. doi: 10.1016/j.bpj.2010.07.049. Biophys J. 2010. PMID: 20923647 Free PMC article.
-
Molecular Dynamics Simulations of Curved Lipid Membranes.Int J Mol Sci. 2022 Jul 22;23(15):8098. doi: 10.3390/ijms23158098. Int J Mol Sci. 2022. PMID: 35897670 Free PMC article. Review.
-
High curvature promotes fusion of lipid membranes: Predictions from continuum elastic theory.Biophys J. 2023 May 16;122(10):1868-1882. doi: 10.1016/j.bpj.2023.04.018. Epub 2023 Apr 18. Biophys J. 2023. PMID: 37077047 Free PMC article.
-
The Role of a Crystallographically Unresolved Cytoplasmic Loop in Stabilizing the Bacterial Membrane Insertase YidC2.Sci Rep. 2019 Oct 8;9(1):14451. doi: 10.1038/s41598-019-51052-9. Sci Rep. 2019. PMID: 31595020 Free PMC article.
References
-
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. - PubMed
-
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. - PubMed
-
- Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. - PMC - PubMed
-
- Sprong H, van der Sluijs P, van Meer G. How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol. 2001;2:504–513. - PubMed
-
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

