Revealing biases inherent in recombination protocols
- PMID: 18001472
- PMCID: PMC2203992
- DOI: 10.1186/1472-6750-7-77
Revealing biases inherent in recombination protocols
Abstract
Background: The recombination of homologous genes is an effective protein engineering tool to evolve proteins. DNA shuffling by gene fragmentation and reassembly has dominated the literature since its first publication, but this fragmentation-based method is labor intensive. Recently, a fragmentation-free PCR based protocol has been published, termed recombination-dependent PCR, which is easy to perform. However, a detailed comparison of both methods is still missing.
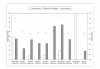
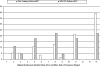
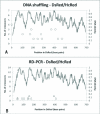
Results: We developed different test systems to compare and reveal biases from DNA shuffling and recombination-dependent PCR (RD-PCR), a StEP-like recombination protocol. An assay based on the reactivation of beta-lactamase was developed to simulate the recombination of point mutations. Both protocols performed similarly here, with slight advantages for RD-PCR. However, clear differences in the performance of the recombination protocols were observed when applied to homologous genes of varying DNA identities. Most importantly, the recombination-dependent PCR showed a less pronounced bias of the crossovers in regions with high sequence identity. We discovered that template variations, including engineered terminal truncations, have significant influence on the position of the crossovers in the recombination-dependent PCR. In comparison, DNA shuffling can produce higher crossover numbers, while the recombination-dependent PCR frequently results in one crossover. Lastly, DNA shuffling and recombination-dependent PCR both produce counter-productive variants such as parental sequences and have chimeras that are over-represented in a library, respectively. Lastly, only RD-PCR yielded chimeras in the low homology situation of GFP/mRFP (45% DNA identity level).
Conclusion: By comparing different recombination scenarios, this study expands on existing recombination knowledge and sheds new light on known biases, which should improve library-creation efforts. It could be shown that the recombination-dependent PCR is an easy to perform alternative to DNA shuffling.
Figures




Similar articles
-
Automating gene library synthesis by structure-based combinatorial protein engineering: examples from plant sesquiterpene synthases.Methods Enzymol. 2012;515:21-42. doi: 10.1016/B978-0-12-394290-6.00002-1. Methods Enzymol. 2012. PMID: 22999168
-
Restriction enzyme-mediated DNA family shuffling.Methods Mol Biol. 2014;1179:175-87. doi: 10.1007/978-1-4939-1053-3_12. Methods Mol Biol. 2014. PMID: 25055778
-
Chimeric gene library construction by a simple and highly versatile method using recombination-dependent exponential amplification.Biotechnol Prog. 2003 Sep-Oct;19(5):1460-7. doi: 10.1021/bp034029t. Biotechnol Prog. 2003. PMID: 14524707
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
-
[Toward creation of proteins by combination of block shuffling and in vitro selection methods].Seikagaku. 2005 Sep;77(9):1165-76. Seikagaku. 2005. PMID: 16241003 Review. Japanese. No abstract available.
Cited by
-
In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the SVZ.Mol Ther. 2018 Jan 3;26(1):304-319. doi: 10.1016/j.ymthe.2017.09.006. Epub 2017 Sep 8. Mol Ther. 2018. PMID: 28988711 Free PMC article.
-
Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy.Biotechnol Biofuels. 2012 Jan 13;5(1):3. doi: 10.1186/1754-6834-5-3. Biotechnol Biofuels. 2012. PMID: 22244361 Free PMC article.
-
The outlook for protein engineering in crop improvement.Plant Physiol. 2008 May;147(1):6-12. doi: 10.1104/pp.108.117929. Plant Physiol. 2008. PMID: 18443101 Free PMC article. Review. No abstract available.
-
Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently.Chem Soc Rev. 2015 Mar 7;44(5):1172-239. doi: 10.1039/c4cs00351a. Chem Soc Rev. 2015. PMID: 25503938 Free PMC article. Review.
-
Amino ester hydrolase from Xanthomonas campestris pv. campestris, ATCC 33913 for enzymatic synthesis of ampicillin.J Mol Catal B Enzym. 2010 Oct;67(1-2):21-28. doi: 10.1016/j.molcatb.2010.06.014. J Mol Catal B Enzym. 2010. PMID: 22087071 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

