Data growth and its impact on the SCOP database: new developments
- PMID: 18000004
- PMCID: PMC2238974
- DOI: 10.1093/nar/gkm993
Data growth and its impact on the SCOP database: new developments
Abstract
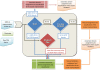
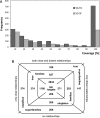
The Structural Classification of Proteins (SCOP) database is a comprehensive ordering of all proteins of known structure, according to their evolutionary and structural relationships. The SCOP hierarchy comprises the following levels: Species, Protein, Family, Superfamily, Fold and Class. While keeping the original classification scheme intact, we have changed the production of SCOP in order to cope with a rapid growth of new structural data and to facilitate the discovery of new protein relationships. We describe ongoing developments and new features implemented in SCOP. A new update protocol supports batch classification of new protein structures by their detected relationships at Family and Superfamily levels in contrast to our previous sequential handling of new structural data by release date. We introduce pre-SCOP, a preview of the SCOP developmental version that enables earlier access to the information on new relationships. We also discuss the impact of worldwide Structural Genomics initiatives, which are producing new protein structures at an increasing rate, on the rates of discovery and growth of protein families and superfamilies. SCOP can be accessed at http://scop.mrc-lmb.cam.ac.uk/scop.
Figures
Similar articles
-
SCOP database in 2002: refinements accommodate structural genomics.Nucleic Acids Res. 2002 Jan 1;30(1):264-7. doi: 10.1093/nar/30.1.264. Nucleic Acids Res. 2002. PMID: 11752311 Free PMC article.
-
SCOP database in 2004: refinements integrate structure and sequence family data.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D226-9. doi: 10.1093/nar/gkh039. Nucleic Acids Res. 2004. PMID: 14681400 Free PMC article.
-
The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures.Nucleic Acids Res. 2020 Jan 8;48(D1):D376-D382. doi: 10.1093/nar/gkz1064. Nucleic Acids Res. 2020. PMID: 31724711 Free PMC article.
-
The SUPERFAMILY database in structural genomics.Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1897-900. doi: 10.1107/s0907444902015160. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393919 Review.
-
Structural classification of proteins and structural genomics: new insights into protein folding and evolution.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Oct 1;66(Pt 10):1190-7. doi: 10.1107/S1744309110007177. Epub 2010 Jul 6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20944210 Free PMC article. Review.
Cited by
-
Vivaldi: visualization and validation of biomacromolecular NMR structures from the PDB.Proteins. 2013 Apr;81(4):583-91. doi: 10.1002/prot.24213. Epub 2013 Jan 15. Proteins. 2013. PMID: 23180575 Free PMC article.
-
DOMMINO: a database of macromolecular interactions.Nucleic Acids Res. 2012 Jan;40(Database issue):D501-6. doi: 10.1093/nar/gkr1128. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135305 Free PMC article.
-
Viral evolution: Primordial cellular origins and late adaptation to parasitism.Mob Genet Elements. 2012 Sep 1;2(5):247-252. doi: 10.4161/mge.22797. Mob Genet Elements. 2012. PMID: 23550145 Free PMC article.
-
Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing.Biol Direct. 2013 Jun 15;8:15. doi: 10.1186/1745-6150-8-15. Biol Direct. 2013. PMID: 23768067 Free PMC article.
-
Ribosomal history reveals origins of modern protein synthesis.PLoS One. 2012;7(3):e32776. doi: 10.1371/journal.pone.0032776. Epub 2012 Mar 12. PLoS One. 2012. PMID: 22427882 Free PMC article.
References
-
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

