Use of DNAzymes for site-specific analysis of ribonucleotide modifications
- PMID: 17998290
- PMCID: PMC2151034
- DOI: 10.1261/rna.742708
Use of DNAzymes for site-specific analysis of ribonucleotide modifications
Abstract
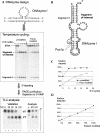
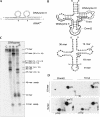
Post-transcriptional ribonucleotide modifications are widespread and abundant processes that have not been analyzed adequately due to the lack of appropriate detection methods. Here, two methods for the analysis of modified nucleotides in RNA are presented that are based on the quantitative and site-specific DNAzyme-mediated cleavage of the target RNA at or near the site of modification. Quantitative RNA cleavage is achieved by cycling the DNAzyme and its RNA substrate through repeated periods of heating and cooling. In a first approach, DNAzyme-directed cleavage directly 5' of the residue in question allows radioactive labeling of the newly freed 5'-OH. After complete enzymatic hydrolysis, the modification status can be assessed by two-dimensional thin layer chromatography. In a second approach, oligoribonucleotide fragments comprising the modification site are excised from the full-length RNA in an endonucleolytic fashion, using a tandem DNAzyme. The excised fragment is isolated by electrophoresis and submitted to further conventional analysis. These results establish DNAzymes as valuable tools for the site-specific and highly sensitive detection of ribonucleotide modifications.
Figures
Similar articles
-
A post-labeling approach for the characterization and quantification of RNA modifications based on site-directed cleavage by DNAzymes.Methods Mol Biol. 2011;718:259-70. doi: 10.1007/978-1-61779-018-8_16. Methods Mol Biol. 2011. PMID: 21370054
-
Target-site selection for the 10-23 DNAzyme.Methods Mol Biol. 2004;252:267-77. doi: 10.1385/1-59259-746-7:267. Methods Mol Biol. 2004. PMID: 15017056
-
Improved RNA cleavage by LNAzyme derivatives of DNAzymes.Biochem Soc Trans. 2004 Feb;32(Pt 1):37-40. doi: 10.1042/bst0320037. Biochem Soc Trans. 2004. PMID: 14748708
-
A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8-17 RNA-cleaving DNAzyme.Chembiochem. 2010 May 3;11(7):866-79. doi: 10.1002/cbic.200900786. Chembiochem. 2010. PMID: 20213779 Review.
-
DNA Catalysis: The Chemical Repertoire of DNAzymes.Molecules. 2015 Nov 20;20(11):20777-804. doi: 10.3390/molecules201119730. Molecules. 2015. PMID: 26610449 Free PMC article. Review.
Cited by
-
SCARPET: site-specific quantification of methylated and nonmethylated adenosines reveals m6A stoichiometry.RNA. 2024 Feb 16;30(3):308-324. doi: 10.1261/rna.079776.123. RNA. 2024. PMID: 38190635 Free PMC article.
-
High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing.Angew Chem Int Ed Engl. 2015 Jan 26;54(5):1587-90. doi: 10.1002/anie.201410647. Epub 2014 Dec 9. Angew Chem Int Ed Engl. 2015. PMID: 25491922 Free PMC article.
-
Genome recoding by tRNA modifications.Open Biol. 2016 Dec;6(12):160287. doi: 10.1098/rsob.160287. Open Biol. 2016. PMID: 27974624 Free PMC article. Review.
-
The RNA modification landscape in human disease.RNA. 2017 Dec;23(12):1754-1769. doi: 10.1261/rna.063503.117. Epub 2017 Aug 30. RNA. 2017. PMID: 28855326 Free PMC article. Review.
-
The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA.RNA Biol. 2017 Sep 2;14(9):1241-1251. doi: 10.1080/15476286.2016.1236170. Epub 2016 Nov 7. RNA Biol. 2017. PMID: 27819523 Free PMC article.
References
-
- Bakin, A., Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. - PubMed
-
- Bakin, A.V., Ofengand, J. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 1998;77:297–309. - PubMed
-
- Brule, H., Grosjean, H., Giege, R., Florentz, C. A pseudoknotted tRNA variant is a substrate for tRNA (cytosine-5)-methyltransferase from Xenopus laevis . Biochimie. 1998;80:977–985. - PubMed
-
- Buchhaupt, M., Peifer, C., Entian, K.D. Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 2007;361:102–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
