An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis
- PMID: 17991777
- PMCID: PMC2084326
- DOI: 10.1073/pnas.0705856104
An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis
Abstract
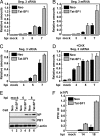
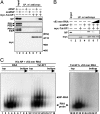
Influenza viruses infect vertebrates, including mammals and birds. Influenza virus reverse-genetics systems facilitate the study of the structure and function of viral factors. In contrast, less is known about host factors involved in the replication process. Here, we developed a replication and transcription system of the negative-strand RNA genome of the influenza virus in Saccharomyces cerevisiae, which depends on viral RNAs, viral RNA polymerases, and nucleoprotein (NP). Disruption of SUB2 encoding an orthologue of human RAF-2p48/UAP56, a previously identified viral RNA synthesis stimulatory host factor, resulted in reduction of the viral RNA synthesis rate. Using a genome-wide set of yeast single-gene deletion strains, we found several host factor candidates affecting viral RNA synthesis. We found that among them, Tat-SF1, a mammalian homologue of yeast CUS2, was a stimulatory host factor in influenza virus RNA synthesis. Tat-SF1 interacted with free NP, but not with NP associated with RNA, and facilitated formation of RNA-NP complexes. These results suggest that Tat-SF1 may function as a molecular chaperone for NP, as does RAF-2p48/UAP56. This system has proven useful for further studies on the mechanism of influenza virus genome replication and transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
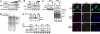
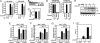
Similar articles
-
Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis.Nat Rev Microbiol. 2016 Aug;14(8):479-93. doi: 10.1038/nrmicro.2016.87. Epub 2016 Jul 11. Nat Rev Microbiol. 2016. PMID: 27396566 Free PMC article. Review.
-
Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis.J Virol. 2001 Feb;75(4):1899-908. doi: 10.1128/JVI.75.4.1899-1908.2001. J Virol. 2001. PMID: 11160689 Free PMC article.
-
Pre-mRNA Processing Factor Prp18 Is a Stimulatory Factor of Influenza Virus RNA Synthesis and Possesses Nucleoprotein Chaperone Activity.J Virol. 2017 Jan 18;91(3):e01398-16. doi: 10.1128/JVI.01398-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27852861 Free PMC article.
-
[Influenza virus replicon and host factors for its replication and transcription].Seikagaku. 2008 Dec;80(12):1128-33. Seikagaku. 2008. PMID: 19172795 Review. Japanese. No abstract available.
-
Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein.J Virol. 2011 Jul;85(13):6197-204. doi: 10.1128/JVI.00277-11. Epub 2011 Apr 20. J Virol. 2011. PMID: 21507964 Free PMC article.
Cited by
-
Identification of putative interactions between swine and human influenza A virus nucleoprotein and human host proteins.Virol J. 2014 Dec 30;11:228. doi: 10.1186/s12985-014-0228-6. Virol J. 2014. PMID: 25547032 Free PMC article.
-
Uncovering the global host cell requirements for influenza virus replication via RNAi screening.Microbes Infect. 2011 May;13(5):516-25. doi: 10.1016/j.micinf.2011.01.012. Epub 2011 Jan 27. Microbes Infect. 2011. PMID: 21276872 Free PMC article. Review.
-
Yeast for virus research.Microb Cell. 2017 Sep 18;4(10):311-330. doi: 10.15698/mic2017.10.592. Microb Cell. 2017. PMID: 29082230 Free PMC article. Review.
-
Assembly and remodeling of viral DNA and RNA replicons regulated by cellular molecular chaperones.Biophys Rev. 2018 Apr;10(2):445-452. doi: 10.1007/s12551-017-0333-z. Epub 2017 Nov 22. Biophys Rev. 2018. PMID: 29170971 Free PMC article. Review.
-
Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis.Nat Rev Microbiol. 2016 Aug;14(8):479-93. doi: 10.1038/nrmicro.2016.87. Epub 2016 Jul 11. Nat Rev Microbiol. 2016. PMID: 27396566 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

