The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells
- PMID: 17962301
- PMCID: PMC2248766
- DOI: 10.1093/nar/gkm888
The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells
Abstract
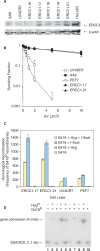
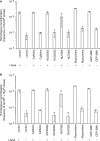
The mammalian ERCC1-XPF endonuclease has a suggested role in the repair of DNA double-strand breaks (DSB) by single-strand annealing (SSA). Here, we investigated the role of ERCC1 in homologous recombination in mammalian cells, and confirm a role of ERCC1 in SSA. Interestingly, we also report an unexpected role for ERCC1 in gene conversion. This provides support that gene conversion in mammalian somatic cells is carried out through synthesis-dependent strand annealing, rather than through a double Holliday Junction mechanism. Moreover, we find low frequencies of SSA and gene conversion in G1-arrested cells, suggesting that SSA is not a frequent DSB repair pathway in G1-arrested mammalian cells, even in the presence of perfect repeats. Furthermore, we find that SSA is not influenced by inhibition of CDK2 (using Roscovitine), ATM (using Caffeine and KU55933), Chk1 (using CEP-3891) or DNA-PK (using NU7026).
Figures

Similar articles
-
Effects of varying gene targeting parameters on processing of recombination intermediates by ERCC1-XPF.DNA Repair (Amst). 2011 Feb 7;10(2):188-98. doi: 10.1016/j.dnarep.2010.10.011. Epub 2010 Nov 30. DNA Repair (Amst). 2011. PMID: 21123118 Free PMC article.
-
Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks.Nucleic Acids Res. 2008 Jul;36(12):4088-98. doi: 10.1093/nar/gkn347. Epub 2008 Jun 6. Nucleic Acids Res. 2008. PMID: 18539610 Free PMC article.
-
The role of nonhomologous DNA end joining, conservative homologous recombination, and single-strand annealing in the cell cycle-dependent repair of DNA double-strand breaks induced by H(2)O(2) in mammalian cells.Radiat Res. 2008 Dec;170(6):784-93. doi: 10.1667/RR1375.1. Radiat Res. 2008. PMID: 19138034
-
Regulation of Single-Strand Annealing and its Role in Genome Maintenance.Trends Genet. 2016 Sep;32(9):566-575. doi: 10.1016/j.tig.2016.06.007. Epub 2016 Jul 19. Trends Genet. 2016. PMID: 27450436 Free PMC article. Review.
-
Interstrand crosslink repair: can XPF-ERCC1 be let off the hook?Trends Genet. 2008 Feb;24(2):70-6. doi: 10.1016/j.tig.2007.11.003. Epub 2008 Jan 14. Trends Genet. 2008. PMID: 18192062 Review.
Cited by
-
Initiation of DNA interstrand cross-link repair in mammalian cells.Environ Mol Mutagen. 2010 Jul;51(6):604-24. doi: 10.1002/em.20559. Environ Mol Mutagen. 2010. PMID: 20658650 Free PMC article. Review.
-
Processing of triplex-directed psoralen DNA interstrand crosslinks by recombination mechanisms.Nucleic Acids Res. 2008 Aug;36(14):4680-8. doi: 10.1093/nar/gkn438. Epub 2008 Jul 15. Nucleic Acids Res. 2008. PMID: 18628293 Free PMC article.
-
Deficiency of nucleotide excision repair is associated with mutational signature observed in cancer.Genome Res. 2019 Jul;29(7):1067-1077. doi: 10.1101/gr.246223.118. Epub 2019 Jun 20. Genome Res. 2019. PMID: 31221724 Free PMC article.
-
Non-B DNA Secondary Structures and Their Resolution by RecQ Helicases.J Nucleic Acids. 2011;2011:724215. doi: 10.4061/2011/724215. Epub 2011 Oct 2. J Nucleic Acids. 2011. PMID: 21977309 Free PMC article.
-
Characterization of CHO XPF mutant UV41: influence of XPF heterozygosity on double-strand break-induced intrachromosomal recombination.DNA Repair (Amst). 2008 Aug 2;7(8):1319-29. doi: 10.1016/j.dnarep.2008.04.012. Epub 2008 Jun 10. DNA Repair (Amst). 2008. PMID: 18547876 Free PMC article.
References
-
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- van Duin M, Vredeveldt G, Mayne LV, Odijk H, Vermeulen W, Klein B, Weeda G, Hoeijmakers JH, Bootsma D, et al. The cloned human DNA excision repair gene ERCC-1 fails to correct xeroderma pigmentosum complementation groups A through I. Mutat. Res. 1989;217:83–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

