MetaLook: a 3D visualisation software for marine ecological genomics
- PMID: 17953757
- PMCID: PMC2134934
- DOI: 10.1186/1471-2105-8-406
MetaLook: a 3D visualisation software for marine ecological genomics
Abstract
Background: Marine ecological genomics can be defined as the application of genomic sciences to understand the structure and function of marine ecosystems. In this field of research, the analysis of genomes and metagenomes of environmental relevance must take into account the corresponding habitat (contextual) data, e.g. water depth, physical and chemical parameters. The creation of specialised software tools and databases is requisite to allow this new kind of integrated analysis.
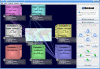
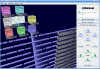
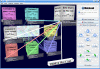
Results: We implemented the MetaLook software for visualisation and analysis of marine ecological genomic and metagenomic data with respect to habitat parameters. MetaLook offers a three-dimensional user interface to interactively visualise DNA sequences on a world map, based on a centralised georeferenced database. The user can define environmental containers to organise the sequences according to different habitat criteria. To find similar sequences, the containers can be queried with either genes from the georeferenced database or user-imported sequences, using the BLAST algorithm. This allows an interactive assessment of the distribution of gene functions in the environment.
Conclusion: MetaLook allows scientists to investigate sequence data in their environmental context and to explore correlations between genes and habitat parameters. This software is a step towards the creation of specialised tools to study constrained distributions and habitat specificity of genes correlated with specific processes. MetaLook is available at: http://www.megx.net/metalook.
Figures

Similar articles
-
MetaMine--a tool to detect and analyse gene patterns in their environmental context.BMC Bioinformatics. 2008 Oct 28;9:459. doi: 10.1186/1471-2105-9-459. BMC Bioinformatics. 2008. PMID: 18957118 Free PMC article.
-
Megx.net: integrated database resource for marine ecological genomics.Nucleic Acids Res. 2010 Jan;38(Database issue):D391-5. doi: 10.1093/nar/gkp918. Epub 2009 Oct 25. Nucleic Acids Res. 2010. PMID: 19858098 Free PMC article.
-
Megx.net--database resources for marine ecological genomics.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D390-3. doi: 10.1093/nar/gkj070. Nucleic Acids Res. 2006. PMID: 16381894 Free PMC article.
-
Ecological and evolutionary genomics of marine photosynthetic organisms.Mol Ecol. 2013 Feb;22(3):867-907. doi: 10.1111/mec.12000. Epub 2012 Sep 18. Mol Ecol. 2013. PMID: 22989289 Review.
-
Visualization techniques for genomic data.Proc IEEE Comput Soc Bioinform Conf. 2002;1:321-6. Proc IEEE Comput Soc Bioinform Conf. 2002. PMID: 15838148 Review.
Cited by
-
A primer on metagenomics.PLoS Comput Biol. 2010 Feb 26;6(2):e1000667. doi: 10.1371/journal.pcbi.1000667. PLoS Comput Biol. 2010. PMID: 20195499 Free PMC article. Review.
-
Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola.PLoS Genet. 2009 Feb;5(2):e1000362. doi: 10.1371/journal.pgen.1000362. Epub 2009 Feb 6. PLoS Genet. 2009. PMID: 19197347 Free PMC article.
-
JCoast - a biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes.BMC Bioinformatics. 2008 Apr 1;9:177. doi: 10.1186/1471-2105-9-177. BMC Bioinformatics. 2008. PMID: 18380896 Free PMC article.
-
MetaMine--a tool to detect and analyse gene patterns in their environmental context.BMC Bioinformatics. 2008 Oct 28;9:459. doi: 10.1186/1471-2105-9-459. BMC Bioinformatics. 2008. PMID: 18957118 Free PMC article.
References
-
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. - DOI - PMC - PubMed
-
- International Nucleotide Sequence Database Collaboration http://www.insdc.org
-
- The Genomic Standards Consortium (GSC) http://darwin.nox.ac.uk/gsc/gcat
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

