Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation
- PMID: 17942535
- PMCID: PMC2168864
- DOI: 10.1128/JVI.01313-07
Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation
Abstract
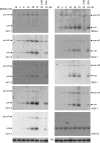
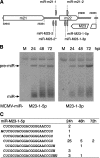
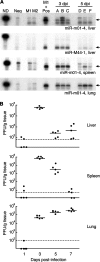
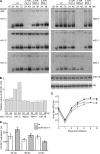
MicroRNAs (miRNAs) are small, noncoding RNA molecules that regulate gene expression at the posttranscriptional level. Originally identified in a variety of organisms ranging from plants to mammals, miRNAs have recently been identified in several viruses. Viral miRNAs may play a role in modulating both viral and host gene expression. Here, we report on the identification and characterization of 18 viral miRNAs from mouse fibroblasts lytically infected with the murine cytomegalovirus (MCMV). The MCMV miRNAs are expressed at early times of infection and are scattered in small clusters throughout the genome with up to four distinct miRNAs processed from a single transcript. No significant homologies to human CMV-encoded miRNAs were found. Remarkably, as soon as 24 h after infection, MCMV miRNAs constituted about 35% of the total miRNA pool, and at 72 h postinfection, this proportion was increased to more than 60%. However, despite the abundance of viral miRNAs during the early phase of infection, the expression of some MCMV miRNAs appeared to be regulated. Hence, for three miRNAs we observed polyuridylation of their 3' end, coupled to subsequent degradation. Individual knockout mutants of two of the most abundant MCMV miRNAs, miR-m01-4 and miR-M44-1, or a double knockout mutant of miR-m21-1 and miR-M23-2, incurred no or only a very mild growth deficit in murine embryonic fibroblasts in vitro.
Figures

Similar articles
-
Manipulation of Viral MicroRNAs as a Potential Antiviral Strategy for the Treatment of Cytomegalovirus Infection.Viruses. 2017 May 19;9(5):118. doi: 10.3390/v9050118. Viruses. 2017. PMID: 28534856 Free PMC article.
-
Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus.J Virol. 2007 Dec;81(24):13761-70. doi: 10.1128/JVI.01290-07. Epub 2007 Oct 10. J Virol. 2007. PMID: 17928340 Free PMC article.
-
Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo.PLoS Pathog. 2012 Feb;8(2):e1002510. doi: 10.1371/journal.ppat.1002510. Epub 2012 Feb 9. PLoS Pathog. 2012. PMID: 22346748 Free PMC article.
-
[Progress on the Function of Herpesvirus-encoded MicroRNAs].Bing Du Xue Bao. 2015 Nov;31(6):704-11. Bing Du Xue Bao. 2015. PMID: 26951018 Review. Chinese.
-
Herpesvirus microRNAs: phenotypes and functions.Curr Opin Virol. 2011 Sep;1(3):211-5. doi: 10.1016/j.coviro.2011.04.003. Curr Opin Virol. 2011. PMID: 21927637 Free PMC article. Review.
Cited by
-
Role of virus-encoded microRNAs in herpesvirus biology.Trends Microbiol. 2009 Dec;17(12):544-53. doi: 10.1016/j.tim.2009.09.002. Epub 2009 Oct 12. Trends Microbiol. 2009. PMID: 19828316 Free PMC article. Review.
-
Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs.RNA. 2010 Aug;16(8):1540-58. doi: 10.1261/rna.1967910. Epub 2010 Jun 21. RNA. 2010. PMID: 20566670 Free PMC article.
-
Noncoding RNPs of viral origin.Cold Spring Harb Perspect Biol. 2011 Mar 1;3(3):a005165. doi: 10.1101/cshperspect.a005165. Cold Spring Harb Perspect Biol. 2011. PMID: 20719877 Free PMC article. Review.
-
MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs.Nature. 2008 Aug 7;454(7205):780-3. doi: 10.1038/nature07103. Epub 2008 Jul 2. Nature. 2008. PMID: 18596690 Free PMC article.
-
Viruses and microRNAs: RISCy interactions with serious consequences.Genes Dev. 2011 Sep 15;25(18):1881-94. doi: 10.1101/gad.17352611. Epub 2011 Sep 6. Genes Dev. 2011. PMID: 21896651 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

