Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex
- PMID: 17939992
- PMCID: PMC2040395
- DOI: 10.1073/pnas.0707266104
Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex
Abstract
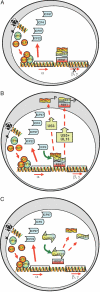
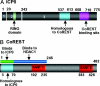
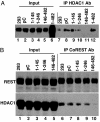
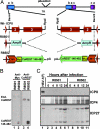
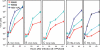
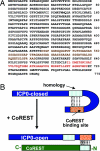
A preeminent phenotype of the infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) is that it acts as a promiscuous transactivator. In most cell lines exposed to DeltaICP0 mutant virus at low ratios of virus per cell infection, alpha genes are expressed but the transition to beta and gamma gene expression does not ensue, but can be enhanced by inhibitors of histone deacetylases (HDACs). Earlier studies have shown that ICP0 interacts with CoREST and displaces HDAC1 from the CoREST-REST-HDAC1/2 complex. HDAC1 and CoREST are then independently translocated to the cytoplasm. Here, we test the hypothesis that ICP0 blocks the silencing of HSV DNA by displacing HDAC1 from the CoREST-REST complex. Specifically, first, mapping studies led us to construct a truncated CoREST (CoREST(146-482)) that in transfected cells displaced HDAC1 from the CoREST-REST complex. Second, we constructed two viruses. In BACs encoding the entire HSV-1, we replaced the gene encoding ICP0 with AmpR to yield a DeltaICP0 mutant R8501. We also replaced ICP0 with CoREST(146-482) to yield recombinant R8502. The yield of R8502 mutant virus in Vero, HEp-2, and human embryonic lung cells exposed to 0.1 pfu of virus per cell was 100-, 10-, and 10-fold higher, respectively, than those of R8501 mutant virus. In Vero cells, the yield of R8502 was identical with that of wild-type virus. We conclude that CoREST(146-482) functionally replaced ICP0 and that, by extension, ICP0 acts to block the silencing of viral DNA by displacing HDAC1/2 from the CoREST-REST complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells.Proc Natl Acad Sci U S A. 2005 May 24;102(21):7571-6. doi: 10.1073/pnas.0502658102. Epub 2005 May 16. Proc Natl Acad Sci U S A. 2005. PMID: 15897453 Free PMC article.
-
The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem.J Virol. 2009 Jan;83(1):181-7. doi: 10.1128/JVI.01940-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945770 Free PMC article.
-
Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1.J Virol. 2009 May;83(9):4376-85. doi: 10.1128/JVI.02515-08. Epub 2009 Feb 4. J Virol. 2009. PMID: 19193804 Free PMC article.
-
The first 30 minutes in the life of a virus: unREST in the nucleus.Cell Cycle. 2005 Aug;4(8):1019-21. doi: 10.4161/cc.4.8.1902. Epub 2005 Aug 7. Cell Cycle. 2005. PMID: 16082207 Review.
-
The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex.J Virol. 2011 Aug;85(15):7474-82. doi: 10.1128/JVI.00180-11. Epub 2011 Mar 30. J Virol. 2011. PMID: 21450817 Free PMC article. Review.
Cited by
-
Role of herpes simplex virus ICP0 in the transactivation of genes introduced by infection or transfection: a reappraisal.J Virol. 2010 May;84(9):4222-8. doi: 10.1128/JVI.02585-09. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164233 Free PMC article.
-
A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses.EMBO J. 2010 Mar 3;29(5):943-55. doi: 10.1038/emboj.2009.400. Epub 2010 Jan 14. EMBO J. 2010. PMID: 20075863 Free PMC article.
-
Depletion of CoREST does not improve the replication of ICP0 null mutant herpes simplex virus type 1.J Virol. 2010 Apr;84(7):3695-8. doi: 10.1128/JVI.00021-10. Epub 2010 Jan 27. J Virol. 2010. PMID: 20106915 Free PMC article.
-
Sequences related to SUMO interaction motifs in herpes simplex virus 1 protein ICP0 act cooperatively to stimulate virus infection.J Virol. 2014 Mar;88(5):2763-74. doi: 10.1128/JVI.03417-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352468 Free PMC article.
-
IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications.PLoS Pathog. 2014 Nov 6;10(11):e1004503. doi: 10.1371/journal.ppat.1004503. eCollection 2014 Nov. PLoS Pathog. 2014. PMID: 25375629 Free PMC article.
References
-
- Roizman B, Knipe DM, Whitley RJ. In: Fields' Virology. 5th Ed. Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. New York: Lippincott Williams & Wilkins; 2007. pp. 2501–2601.
-
- Wysocka J, Herr W. Trends Biochem Sci. 2003;28:294–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

