Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae
- PMID: 17923688
- PMCID: PMC2169175
- DOI: 10.1128/MCB.01543-07
Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae
Abstract
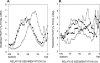
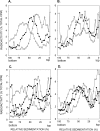
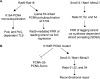
In Saccharomyces cerevisiae, postreplication repair (PRR) of UV-damaged DNA occurs by a Rad6-Rad18- and an Mms2-Ubc13-Rad5-dependent pathway or by a Rad52-dependent pathway. The Rad5 DNA helicase activity is specialized for promoting replication fork regression and template switching; previously, we suggested a role for the Rad5-dependent PRR pathway when the lesion is located on the leading strand and a role for the Rad52 pathway when the lesion is located on the lagging strand. In this study, we present evidence for the requirement of Nse1, a subunit of the Smc5-Smc6 complex, in Rad52-dependent PRR, and our genetic analyses suggest a role for the Nse1 and Mms21 E3 ligase activities associated with this complex in this repair mode. We discuss the possible ways by which the Smc5-Smc6 complex, including its associated ubiquitin ligase and SUMO ligase activities, might contribute to the Rad52-dependent nonrecombinational and recombinational modes of PRR.
Figures




Similar articles
-
Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Nov;27(21):7758-64. doi: 10.1128/MCB.01331-07. Epub 2007 Sep 4. Mol Cell Biol. 2007. PMID: 17785441 Free PMC article.
-
Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Apr;22(7):2419-26. doi: 10.1128/MCB.22.7.2419-2426.2002. Mol Cell Biol. 2002. PMID: 11884624 Free PMC article.
-
Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae.Mol Cell Biol. 2004 May;24(10):4267-74. doi: 10.1128/MCB.24.10.4267-4274.2004. Mol Cell Biol. 2004. PMID: 15121847 Free PMC article.
-
Regulation of rDNA stability by sumoylation.DNA Repair (Amst). 2009 Apr 5;8(4):507-16. doi: 10.1016/j.dnarep.2009.01.015. Epub 2009 Mar 3. DNA Repair (Amst). 2009. PMID: 19261548 Review.
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
Cited by
-
BLM SUMOylation regulates ssDNA accumulation at stalled replication forks.Front Genet. 2013 Sep 4;4:167. doi: 10.3389/fgene.2013.00167. eCollection 2013. Front Genet. 2013. PMID: 24027577 Free PMC article.
-
SUMOylation regulates the SNF1 protein kinase.Proc Natl Acad Sci U S A. 2013 Oct 22;110(43):17432-7. doi: 10.1073/pnas.1304839110. Epub 2013 Oct 9. Proc Natl Acad Sci U S A. 2013. PMID: 24108357 Free PMC article.
-
The unnamed complex: what do we know about Smc5-Smc6?Chromosome Res. 2009;17(2):251-63. doi: 10.1007/s10577-008-9016-8. Chromosome Res. 2009. PMID: 19308705 Review.
-
Loss of Caenorhabditis elegans BRCA1 promotes genome stability during replication in smc-5 mutants.Genetics. 2014 Apr;196(4):985-99. doi: 10.1534/genetics.113.158295. Epub 2014 Jan 14. Genetics. 2014. PMID: 24424777 Free PMC article.
-
Scaffolding for Repair: Understanding Molecular Functions of the SMC5/6 Complex.Genes (Basel). 2018 Jan 12;9(1):36. doi: 10.3390/genes9010036. Genes (Basel). 2018. PMID: 29329249 Free PMC article. Review.
References
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
-
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. - PubMed
-
- Cordeiro-Stone, M., A. M. Makhov, L. S. Zaritskaya, and J. D. Griffith. 1999. Analysis of DNA replication forks encountering a pyrmidine dimer in the template to the leading strand. J. Mol. Biol. 289:1207-1218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
