Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases
- PMID: 17908815
- PMCID: PMC2168330
- DOI: 10.1128/IAI.01020-07
Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases
Abstract
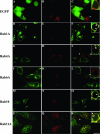
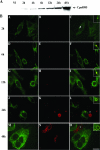
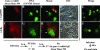
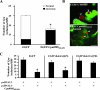
Chlamydiae are intracellular bacteria that develop within a membrane-bound vacuole called an inclusion. To ensure that the inclusion is a safe niche for chlamydial replication, chlamydiae exploit a number of host cell processes, including membrane-trafficking pathways. Recently, several Rab GTPases were found to associate with the inclusions of various chlamydial species. Here we report that Cpn0585, a Chlamydia pneumoniae inclusion membrane protein (Inc), interacts with multiple Rab GTPases. The results from yeast two-hybrid experiments revealed that an amino-terminally truncated form of Cpn0585 (Cpn0585(102-651)) interacts with Rab1, Rab10, and Rab11 but not with Rab4 or Rab6. Cpn0585-Rab GTPase interactions are direct and GTP dependent as shown in glutathione S-transferase pull-down assays using native and recombinant Cpn0585. In C. pneumoniae-infected HEp-2 cells transfected with enhanced green fluorescent protein (EGFP)-tagged Rab GTPases, the colocalization with Cpn0585 at the inclusion membrane was partial for EGFP-Rab1 and EGFP-Rab10, but extensive for wild-type EGFP-Rab11A and the constitutively active GTPase-deficient EGFP-Rab11AQ70L. Moreover, Cpn0585 colocalized with EGFP-Rab11AQ70L as early as 2 h postinfection. Upon delivery into live C. pneumoniae-infected cells, Cpn0585(628-651)-specific antibodies bound to the inclusion membrane, demonstrating that the Rab GTPase-interacting domain of Cpn0585 faces the host cell cytosol. Finally, ectopic expression of Cpn0585(102-651) partially inhibited the development of C. pneumoniae inclusions in EGFP. but not in EGFP-Rab11AQ70L-expressing HEp-2 cells. Collectively, these data suggest that Cpn0585 is involved in the recruitment of Rab GTPases to the inclusion membrane and that interfering with this function may adversely impact the fitness of the C. pneumoniae inclusion for chlamydial replication.
Figures



Similar articles
-
Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner.Infect Immun. 2003 Oct;71(10):5855-70. doi: 10.1128/IAI.71.10.5855-5870.2003. Infect Immun. 2003. PMID: 14500507 Free PMC article.
-
Localization of the hypothetical protein Cpn0585 in the inclusion membrane of Chlamydia pneumoniae-infected cells.Microb Pathog. 2007 Feb-Mar;42(2-3):111-6. doi: 10.1016/j.micpath.2006.11.006. Epub 2007 Jan 22. Microb Pathog. 2007. PMID: 17236746 Free PMC article.
-
The Rab6 effector Bicaudal D1 associates with Chlamydia trachomatis inclusions in a biovar-specific manner.Infect Immun. 2007 Feb;75(2):781-91. doi: 10.1128/IAI.01447-06. Epub 2006 Nov 13. Infect Immun. 2007. PMID: 17101644 Free PMC article.
-
[Effector proteins of Clamidia].Mol Biol (Mosk). 2009 Nov-Dec;43(6):963-83. Mol Biol (Mosk). 2009. PMID: 20088373 Review. Russian.
-
How can mammalian Rab small GTPases be comprehensively analyzed?: Development of new tools to comprehensively analyze mammalian Rabs in membrane traffic.Histol Histopathol. 2010 Nov;25(11):1473-80. doi: 10.14670/HH-25.1473. Histol Histopathol. 2010. PMID: 20865669 Review.
Cited by
-
Recombinant Cpn 0810 stimulates proinflammatory cytokine expression and apoptosis in human monocytes.Exp Ther Med. 2015 Feb;9(2):459-463. doi: 10.3892/etm.2014.2111. Epub 2014 Dec 5. Exp Ther Med. 2015. PMID: 25574216 Free PMC article.
-
Functional interaction between type III-secreted protein IncA of Chlamydophila psittaci and human G3BP1.PLoS One. 2011 Jan 31;6(1):e16692. doi: 10.1371/journal.pone.0016692. PLoS One. 2011. PMID: 21304914 Free PMC article.
-
Hijacking host cell vesicular transport: New insights into the nutrient acquisition mechanism of Chlamydia.Virulence. 2024 Dec;15(1):2351234. doi: 10.1080/21505594.2024.2351234. Epub 2024 May 21. Virulence. 2024. PMID: 38773735 Free PMC article. Review.
-
Chlamydial intracellular survival strategies.Cold Spring Harb Perspect Med. 2013 May 1;3(5):a010256. doi: 10.1101/cshperspect.a010256. Cold Spring Harb Perspect Med. 2013. PMID: 23637308 Free PMC article. Review.
-
The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes.Cell Microbiol. 2010 Sep 1;12(9):1292-307. doi: 10.1111/j.1462-5822.2010.01468.x. Epub 2010 Mar 25. Cell Microbiol. 2010. PMID: 20345488 Free PMC article.
References
-
- Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2:35-47. - PubMed
-
- Beatty, W. L. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119(Pt. 2):350-359. - PubMed
-
- Belland, R., D. M. Ojcius, and G. I. Byrne. 2004. Chlamydia. Nat. Rev. Microbiol. 2:530-531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

