Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis
- PMID: 17891140
- PMCID: PMC2254521
- DOI: 10.1038/ncb1634
Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis
Abstract
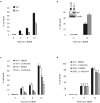
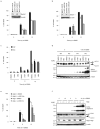
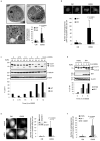
Autophagy is an evolutionarily conserved 'self-eating' process. Although the genes essential for autophagy (named Atg) have been identified in yeast, the molecular mechanism of how Atg proteins control autophagosome formation in mammalian cells remains to be elucidated. Here, we demonstrate that Bif-1 (also known as Endophilin B1) interacts with Beclin 1 through ultraviolet irradiation resistance-associated gene (UVRAG) and functions as a positive mediator of the class III PI(3) kinase (PI(3)KC3). In response to nutrient deprivation, Bif-1 localizes to autophagosomes where it colocalizes with Atg5, as well as microtubule-associated protein light chain 3 (LC3). Furthermore, loss of Bif-1 suppresses autophagosome formation. Although the SH3 domain of Bif-1 is sufficient for binding to UVRAG, both the BAR and SH3 domains are required for Bif-1 to activate PI(3)KC3 and induce autophagosome formation. We also observed that Bif-1 ablation prolongs cell survival under starvation conditions. Moreover, knockout of Bif-1 significantly enhances the development of spontaneous tumours in mice. These findings suggest that Bif-1 joins the UVRAG-Beclin 1 complex as a potential activator of autophagy and tumour suppressor.
Figures


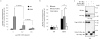

Comment in
-
Shaping membranes into autophagosomes.Nat Cell Biol. 2007 Oct;9(10):1125-7. doi: 10.1038/ncb1007-1125. Nat Cell Biol. 2007. PMID: 17909525 No abstract available.
Similar articles
-
BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1.Autophagy. 2008 Jan;4(1):121-4. doi: 10.4161/auto.5265. Epub 2007 Nov 5. Autophagy. 2008. PMID: 18032918
-
Bif-1/endophilin B1: a candidate for crescent driving force in autophagy.Cell Death Differ. 2009 Jul;16(7):947-55. doi: 10.1038/cdd.2009.19. Epub 2009 Mar 6. Cell Death Differ. 2009. PMID: 19265852 Free PMC article. Review.
-
Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.Mol Biol Cell. 2008 Dec;19(12):5360-72. doi: 10.1091/mbc.e08-01-0080. Epub 2008 Oct 8. Mol Biol Cell. 2008. PMID: 18843052 Free PMC article.
-
A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic.Exp Cell Res. 2010 Dec 10;316(20):3368-78. doi: 10.1016/j.yexcr.2010.07.008. Epub 2010 Jul 17. Exp Cell Res. 2010. PMID: 20643123
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
-
ATG16L1: A multifunctional susceptibility factor in Crohn disease.Autophagy. 2015 Apr 3;11(4):585-94. doi: 10.1080/15548627.2015.1017187. Autophagy. 2015. PMID: 25906181 Free PMC article. Review.
-
Low expression of Beclin 1 and elevated expression of HIF-1α refine distant metastasis risk and predict poor prognosis of ER-positive, HER2-negative breast cancer.Med Oncol. 2013 Mar;30(1):355. doi: 10.1007/s12032-012-0355-0. Epub 2013 Feb 14. Med Oncol. 2013. PMID: 23408367
-
Focusing on cyclin-dependent kinases 5: A potential target for neurological disorders.Front Mol Neurosci. 2022 Nov 10;15:1030639. doi: 10.3389/fnmol.2022.1030639. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36438186 Free PMC article. Review.
-
Inhibition of autophagic turnover in β-cells by fatty acids and glucose leads to apoptotic cell death.J Biol Chem. 2015 Mar 6;290(10):6071-85. doi: 10.1074/jbc.M114.605345. Epub 2014 Dec 29. J Biol Chem. 2015. PMID: 25548282 Free PMC article.
-
The role of autophagy induced by tumor microenvironment in different cells and stages of cancer.Cell Biosci. 2015 Mar 28;5:14. doi: 10.1186/s13578-015-0005-2. eCollection 2015. Cell Biosci. 2015. PMID: 25844158 Free PMC article.
References
-
- Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–8. - PubMed
-
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. - PubMed
-
- Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. - PubMed
-
- Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

