A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase
- PMID: 17884090
- PMCID: PMC2083577
- DOI: 10.1016/j.jmb.2007.08.040
A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase
Abstract
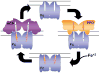
Ferrochelatase (protoheme ferrolyase, EC 4.99.1.1) is the terminal enzyme in heme biosynthesis and catalyzes the insertion of ferrous iron into protoporphyrin IX to form protoheme IX (heme). Due to the many critical roles of heme, synthesis of heme is required by the vast majority of organisms. Despite significant investigation of both the microbial and eukaryotic enzyme, details of metal chelation remain unidentified. Here we present the first structure of the wild-type human enzyme, a lead-inhibited intermediate of the wild-type enzyme with bound metallated porphyrin macrocycle, the product bound form of the enzyme, and a higher resolution model for the substrate-bound form of the E343K variant. These data paint a picture of an enzyme that undergoes significant changes in secondary structure during the catalytic cycle. The role that these structural alterations play in overall catalysis and potential protein-protein interactions with other proteins, as well as the possible molecular basis for these changes, is discussed. The atomic details and structural rearrangements presented herein significantly advance our understanding of the substrate binding mode of ferrochelatase and reveal new conformational changes in a structurally conserved pi-helix that is predicted to have a central role in product release.
Figures

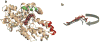
Similar articles
-
Product release rather than chelation determines metal specificity for ferrochelatase.J Mol Biol. 2009 Oct 23;393(2):308-19. doi: 10.1016/j.jmb.2009.08.042. Epub 2009 Aug 22. J Mol Biol. 2009. PMID: 19703464 Free PMC article.
-
Substrate interactions with human ferrochelatase.Proc Natl Acad Sci U S A. 2007 Feb 6;104(6):1789-93. doi: 10.1073/pnas.0606144104. Epub 2007 Jan 29. Proc Natl Acad Sci U S A. 2007. PMID: 17261801 Free PMC article.
-
Ferrochelatase π-helix: Implications from examining the role of the conserved π-helix glutamates in porphyrin metalation and product release.Arch Biochem Biophys. 2018 Apr 15;644:37-46. doi: 10.1016/j.abb.2018.02.015. Epub 2018 Feb 23. Arch Biochem Biophys. 2018. PMID: 29481781 Free PMC article.
-
Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters.Cell Mol Life Sci. 2000 Dec;57(13-14):1909-26. doi: 10.1007/pl00000672. Cell Mol Life Sci. 2000. PMID: 11215517 Free PMC article. Review.
-
Ferrochelatase.Int J Biochem Cell Biol. 1999 Oct;31(10):995-1000. doi: 10.1016/s1357-2725(99)00066-7. Int J Biochem Cell Biol. 1999. PMID: 10582332 Review.
Cited by
-
Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis.Blood. 2010 Jul 29;116(4):628-30. doi: 10.1182/blood-2009-12-259614. Epub 2010 Apr 28. Blood. 2010. PMID: 20427704 Free PMC article.
-
Structure and function of enzymes in heme biosynthesis.Protein Sci. 2010 Jun;19(6):1137-61. doi: 10.1002/pro.405. Protein Sci. 2010. PMID: 20506125 Free PMC article. Review.
-
New Avenues of Heme Synthesis Regulation.Int J Mol Sci. 2022 Jul 5;23(13):7467. doi: 10.3390/ijms23137467. Int J Mol Sci. 2022. PMID: 35806474 Free PMC article. Review.
-
Dimeric ferrochelatase bridges ABCB7 and ABCB10 homodimers in an architecturally defined molecular complex required for heme biosynthesis.Haematologica. 2019 Sep;104(9):1756-1767. doi: 10.3324/haematol.2018.214320. Epub 2019 Feb 14. Haematologica. 2019. PMID: 30765471 Free PMC article.
-
Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product.Microbiol Mol Biol Rev. 2017 Jan 25;81(1):e00048-16. doi: 10.1128/MMBR.00048-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2017. PMID: 28123057 Free PMC article. Review.
References
-
- Dailey HA. Ferrochelatase. In: Hausinger RP, Eichhorn GL, Marzilli LG, editors. Mechanisms of Metallocenter Assembly. VCH Publisher, Inc; New York: 1996. pp. 77–98.
-
- Dunn LL, Rahmanto YS, Richardson DR. Iron uptake and metabolism in the new millennium. Trends Cell Biol. 2007;17:93–100. - PubMed
-
- Hou S, Reynolds MF, Horrigan FT, Heinemann SH, Hoshi T. Reversible binding of heme to proteins in cellular signal transduction. Acc Chem Res. 2006;39:918–24. - PubMed
-
- Mense SM, Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–92. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

