The structure of bacterial ParM filaments
- PMID: 17873883
- PMCID: PMC3541950
- DOI: 10.1038/nsmb1300
The structure of bacterial ParM filaments
Abstract
Bacterial ParM is a homolog of eukaryotic actin and is involved in moving plasmids so that they segregate properly during cell division. Using cryo-EM and three-dimensional reconstruction, we show that ParM filaments have a different structure from F-actin, with very different subunit-subunit interfaces. These interfaces result in the helical handedness of the ParM filament being opposite to that of F-actin. Like F-actin, ParM filaments have a variable twist, and we show that this involves domain-domain rotations within the ParM subunit. The present results yield new insights into polymorphisms within F-actin, as well as the evolution of polymer families.
Figures


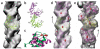
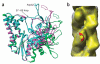

Similar articles
-
Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles.Nature. 2015 Jul 2;523(7558):106-10. doi: 10.1038/nature14356. Epub 2015 Apr 27. Nature. 2015. PMID: 25915019 Free PMC article.
-
Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation.Science. 2009 Jan 23;323(5913):509-12. doi: 10.1126/science.1164346. Epub 2008 Dec 18. Science. 2009. PMID: 19095899
-
F-actin-like filaments formed by plasmid segregation protein ParM.EMBO J. 2002 Dec 16;21(24):6935-43. doi: 10.1093/emboj/cdf672. EMBO J. 2002. PMID: 12486014 Free PMC article.
-
A tale of two polymers: new insights into helical filaments.Nat Rev Mol Cell Biol. 2003 Aug;4(8):621-30. doi: 10.1038/nrm1176. Nat Rev Mol Cell Biol. 2003. PMID: 12923524 Review.
-
Actin's prokaryotic homologs.Curr Opin Struct Biol. 2003 Apr;13(2):244-8. doi: 10.1016/s0959-440x(03)00027-7. Curr Opin Struct Biol. 2003. PMID: 12727519 Review.
Cited by
-
Archaeal actin from a hyperthermophile forms a single-stranded filament.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):9340-5. doi: 10.1073/pnas.1509069112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124094 Free PMC article.
-
Structural plasticity in actin and tubulin polymer dynamics.Science. 2009 Aug 21;325(5943):960-3. doi: 10.1126/science.1168823. Science. 2009. PMID: 19696342 Free PMC article. Review.
-
When cytoskeletal worlds collide.Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19609-10. doi: 10.1073/pnas.1014665107. Epub 2010 Nov 8. Proc Natl Acad Sci U S A. 2010. PMID: 21059902 Free PMC article. No abstract available.
-
Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A.Mol Microbiol. 2009 Aug;73(4):534-52. doi: 10.1111/j.1365-2958.2009.06771.x. Epub 2009 Jul 7. Mol Microbiol. 2009. PMID: 19602153 Free PMC article.
-
ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA.Proc Natl Acad Sci U S A. 2010 Mar 9;107(10):4590-5. doi: 10.1073/pnas.0913060107. Epub 2010 Feb 22. Proc Natl Acad Sci U S A. 2010. PMID: 20176965 Free PMC article.
References
-
- van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. - PubMed
-
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. - PubMed
-
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

