DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication
- PMID: 17855521
- PMCID: PMC2168844
- DOI: 10.1128/JVI.01517-07
DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication
Abstract
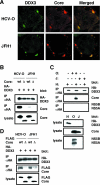
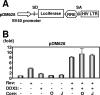
DDX3, a DEAD-box RNA helicase, binds to the hepatitis C virus (HCV) core protein. However, the role(s) of DDX3 in HCV replication is still not understood. Here we demonstrate that the accumulation of both genome-length HCV RNA (HCV-O, genotype 1b) and its replicon RNA were significantly suppressed in HuH-7-derived cells expressing short hairpin RNA targeted to DDX3 by lentivirus vector transduction. As well, RNA replication of JFH1 (genotype 2a) and release of the core into the culture supernatants were suppressed in DDX3 knockdown cells after inoculation of the cell culture-generated HCVcc. Thus, DDX3 is required for HCV RNA replication.
Figures

Similar articles
-
Hepatitis C virus core protein interacts with a human DEAD box protein DDX3.Virology. 1999 May 10;257(2):330-40. doi: 10.1006/viro.1999.9659. Virology. 1999. PMID: 10329544
-
DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control.Oncogene. 2006 Mar 30;25(14):1991-2003. doi: 10.1038/sj.onc.1209239. Oncogene. 2006. PMID: 16301996
-
Cell culture-adaptive NS3 mutations required for the robust replication of genome-length hepatitis C virus RNA.Virus Res. 2007 Apr;125(1):88-97. doi: 10.1016/j.virusres.2006.12.011. Epub 2007 Jan 18. Virus Res. 2007. PMID: 17239465
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
-
In vitro replication models for the hepatitis C virus.J Viral Hepat. 2007 Jan;14(1):2-10. doi: 10.1111/j.1365-2893.2006.00807.x. J Viral Hepat. 2007. PMID: 17212638 Review.
Cited by
-
EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication.J Virol. 2013 Jun;87(12):6625-34. doi: 10.1128/JVI.01006-12. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552423 Free PMC article.
-
Caspase-Dependent Cleavage of DDX21 Suppresses Host Innate Immunity.mBio. 2021 Jun 29;12(3):e0100521. doi: 10.1128/mBio.01005-21. Epub 2021 Jun 14. mBio. 2021. PMID: 34125604 Free PMC article.
-
Cytoplasmic dsRNA induces the expression of OCT3/4 and NANOG mRNAs in differentiated human cells.J Biol Chem. 2019 Dec 13;294(50):18969-18979. doi: 10.1074/jbc.RA119.009783. Epub 2019 Oct 15. J Biol Chem. 2019. PMID: 31615841 Free PMC article.
-
P bodies, stress granules, and viral life cycles.Cell Host Microbe. 2008 Apr 17;3(4):206-12. doi: 10.1016/j.chom.2008.03.004. Cell Host Microbe. 2008. PMID: 18407064 Free PMC article. Review.
-
Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation.EMBO J. 2008 Aug 6;27(15):2147-57. doi: 10.1038/emboj.2008.143. Epub 2008 Jul 17. EMBO J. 2008. PMID: 18636090 Free PMC article.
References
-
- Ariumi, Y., A. Kaida, M. Hatanaka, and K. Shimotohno. 2001. Functional cross-talk of HIV-1 Tat with p53 through its C-terminal domain. Biochem. Biophys. Res. Commun. 287:556-561. - PubMed
-
- Ariumi, Y., T. Ego, A. Kaida, M. Matsumoto, P. P. Pandolfi, and K. Shimotohno. 2003. Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene 22:1611-1619. - PubMed
-
- Bridge, A. J., S. Pebernard, A. Ducraux, A.-L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

