Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity
- PMID: 17804645
- PMCID: PMC2040088
- DOI: 10.1261/rna.553807
Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity
Abstract
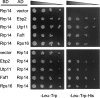
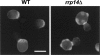
We previously cloned RRP14/YKL082c, whose product exhibits two-hybrid interaction with Ebp2p, a regulatory factor of assembly of 60S ribosomal subunits. Depletion of Rrp14p results in shortage of 60S ribosomal subunits and retardation of processing from 27S pre-rRNA to 25S rRNA. Furthermore, 35S pre-rRNA synthesis appears to decline in Rrp14p-depleted cells. Rrp14p interacts with regulatory factors of 60S subunit assembly and also with Utp11p and Faf1p, which are regulatory factors required for assembly of 40S ribosomal subunits. We propose that Rrp14p is involved in ribosome synthesis from the beginning of 35S pre-rRNA synthesis to assembly of the 60S ribosomal subunit. Disruption of RRP14 causes an extremely slow growth rate of the cell, a severe defect in ribosome synthesis, and a depolarized localization of cortical actin patches throughout the cell cycle. These results suggest that Rrp14p has dual functions in ribosome synthesis and polarized cell growth.
Figures


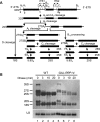
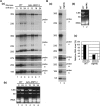
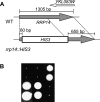
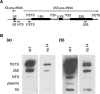

Similar articles
-
Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.Mol Microbiol. 2004 Apr;52(1):141-58. doi: 10.1111/j.1365-2958.2003.03973.x. Mol Microbiol. 2004. PMID: 15049817
-
Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis.Nucleic Acids Res. 2007;35(4):1354-66. doi: 10.1093/nar/gkl824. Epub 2007 Feb 1. Nucleic Acids Res. 2007. PMID: 17272295 Free PMC article.
-
Role of the yeast ribosomal protein L16 in ribosome biogenesis.FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
Cited by
-
Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres.EMBO J. 2011 Aug 5;30(18):3799-811. doi: 10.1038/emboj.2011.267. EMBO J. 2011. PMID: 21822217 Free PMC article.
-
Ribosomal protein L14 contributes to the early assembly of 60S ribosomal subunits in Saccharomyces cerevisiae.Nucleic Acids Res. 2018 May 18;46(9):4715-4732. doi: 10.1093/nar/gky123. Nucleic Acids Res. 2018. PMID: 29788267 Free PMC article.
-
Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast.J Biol Chem. 2014 Aug 15;289(33):22692-22703. doi: 10.1074/jbc.M114.584490. Epub 2014 Jul 2. J Biol Chem. 2014. PMID: 24990943 Free PMC article.
-
Analysis of cell cycle parameters during the transition from unhindered growth to ribosomal and translational stress conditions.PLoS One. 2017 Oct 13;12(10):e0186494. doi: 10.1371/journal.pone.0186494. eCollection 2017. PLoS One. 2017. PMID: 29028845 Free PMC article.
-
Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes.Mol Biol Cell. 2013 Dec;24(23):3620-33. doi: 10.1091/mbc.E13-02-0097. Epub 2013 Oct 9. Mol Biol Cell. 2013. PMID: 24109599 Free PMC article.
References
-
- Bassler, J., Grandi, P., Gadal, O., Lebmann, T., Petfalski, E., Tollervey, D., Lechner, J., Hurt, E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. - PubMed
-
- Cabib, E., Roh, D.H., Schmidt, M., Crotti, L.B., Varma, A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 2001;276:19679–19682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
