Effects of identity minimization on Moloney murine leukemia virus template recognition and frequent tertiary template-directed insertions during nonhomologous recombination
- PMID: 17804514
- PMCID: PMC2168973
- DOI: 10.1128/JVI.01591-07
Effects of identity minimization on Moloney murine leukemia virus template recognition and frequent tertiary template-directed insertions during nonhomologous recombination
Abstract
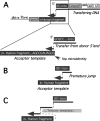
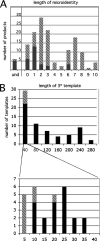
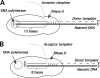
Homology requirements for Moloney murine leukemia virus recombination were addressed in this study by monitoring titer defects observed when acceptor/donor template identity lengths were systematically reduced. Recombination acceptors with at least 16 contiguous bases of donor template identity were recognized as efficiently as longer acceptors. In contrast, a sharp 1-log titer drop was observed for an acceptor of only 15 bases long, with an additional 1-log titer decline for an 8-base acceptor and further decreases for shorter acceptors. Eighty-three independent nonhomologous recombination products were sequenced to examine recombination template selection in the absence of significant sequence identity. These replication products contained a total of 152 nonhomologous crossover junctions. Forced copy choice models predict that forced nonhomologous recombination should result in DNA synthesis to the donor template's 5' end, followed by microidentity-guided acceptor template selection. However, only a single product displayed this structure. The majority of examined nonhomologous recombination products contained junction-associated sequence insertions. Most insertions resulted from the use of one or more tertiary templates, recognizable as discontiguous portions of viral or host RNA or minus-strand DNA. The donor/acceptor template microidentity evident at most crossovers reconfirmed the remarkable capability of the reverse transcription machinery to recognize short regions of sequence identity. These results demonstrate that recruitment of discontiguous host or viral sequences is a common way for retroviruses to resolve nonhomologous recombination junctions and provide experimental support for the role of splinting templates in the generation of retroviral insertions.
Figures






Similar articles
-
Cis-acting elements required for strong stop acceptor template selection during Moloney murine leukemia virus reverse transcription.J Mol Biol. 1998 Aug 7;281(1):1-15. doi: 10.1006/jmbi.1998.1929. J Mol Biol. 1998. PMID: 9680471
-
Effects of limiting homology at the site of intermolecular recombinogenic template switching during Moloney murine leukemia virus replication.J Virol. 2001 Dec;75(23):11263-74. doi: 10.1128/JVI.75.23.11263-11274.2001. J Virol. 2001. PMID: 11689606 Free PMC article.
-
Mismatch extension during strong stop strand transfer and minimal homology requirements for replicative template switching during Moloney murine leukemia virus replication.J Mol Biol. 2003 Jul 18;330(4):657-74. doi: 10.1016/s0022-2836(03)00597-7. J Mol Biol. 2003. PMID: 12850138 Free PMC article.
-
RNA recombination in brome mosaic virus, a model plus strand RNA virus.Acta Biochim Pol. 1998;45(4):847-68. Acta Biochim Pol. 1998. PMID: 10397334 Review.
-
RNA recombination in animal and plant viruses.Microbiol Rev. 1992 Mar;56(1):61-79. doi: 10.1128/mr.56.1.61-79.1992. Microbiol Rev. 1992. PMID: 1579113 Free PMC article. Review.
Cited by
-
The remarkable frequency of human immunodeficiency virus type 1 genetic recombination.Microbiol Mol Biol Rev. 2009 Sep;73(3):451-80, Table of Contents. doi: 10.1128/MMBR.00012-09. Microbiol Mol Biol Rev. 2009. PMID: 19721086 Free PMC article. Review.
-
The Host RNAs in Retroviral Particles.Viruses. 2016 Aug 19;8(8):235. doi: 10.3390/v8080235. Viruses. 2016. PMID: 27548206 Free PMC article. Review.
-
Pseudodiploid genome organization AIDS full-length human immunodeficiency virus type 1 DNA synthesis.J Virol. 2008 Mar;82(5):2376-84. doi: 10.1128/JVI.02100-07. Epub 2007 Dec 19. J Virol. 2008. PMID: 18094172 Free PMC article.
-
Analysis of HIV-1 intersubtype recombination breakpoints suggests region with high pairing probability may be a more fundamental factor than sequence similarity affecting HIV-1 recombination.Virol J. 2016 Sep 21;13(1):156. doi: 10.1186/s12985-016-0616-1. Virol J. 2016. PMID: 27655081 Free PMC article.
-
Resolution of Specific Nucleotide Mismatches by Wild-Type and AZT-Resistant Reverse Transcriptases during HIV-1 Replication.J Mol Biol. 2016 Jun 5;428(11):2275-2288. doi: 10.1016/j.jmb.2016.04.005. Epub 2016 Apr 10. J Mol Biol. 2016. PMID: 27075671 Free PMC article.
References
-
- An, W., and A. Telesnitsky. 2002. HIV-1 genetic recombination: experimental approaches and observations. AIDS Rev. 4:195-212. - PubMed
-
- Ben-Artzi, H., E. Zeelon, B. Amit, A. Wortzel, M. Gorecki, and A. Panet. 1993. RNase H activity of reverse transcriptases on substrates derived from the 5′ end of retroviral genome. J. Biol. Chem. 268:16465-16471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

