A numerical approach to ion channel modelling using whole-cell voltage-clamp recordings and a genetic algorithm
- PMID: 17784781
- PMCID: PMC1963494
- DOI: 10.1371/journal.pcbi.0030169
A numerical approach to ion channel modelling using whole-cell voltage-clamp recordings and a genetic algorithm
Abstract
The activity of trans-membrane proteins such as ion channels is the essence of neuronal transmission. The currently most accurate method for determining ion channel kinetic mechanisms is single-channel recording and analysis. Yet, the limitations and complexities in interpreting single-channel recordings discourage many physiologists from using them. Here we show that a genetic search algorithm in combination with a gradient descent algorithm can be used to fit whole-cell voltage-clamp data to kinetic models with a high degree of accuracy. Previously, ion channel stimulation traces were analyzed one at a time, the results of these analyses being combined to produce a picture of channel kinetics. Here the entire set of traces from all stimulation protocols are analysed simultaneously. The algorithm was initially tested on simulated current traces produced by several Hodgkin-Huxley-like and Markov chain models of voltage-gated potassium and sodium channels. Currents were also produced by simulating levels of noise expected from actual patch recordings. Finally, the algorithm was used for finding the kinetic parameters of several voltage-gated sodium and potassium channels models by matching its results to data recorded from layer 5 pyramidal neurons of the rat cortex in the nucleated outside-out patch configuration. The minimization scheme gives electrophysiologists a tool for reproducing and simulating voltage-gated ion channel kinetics at the cellular level.
Conflict of interest statement
Figures
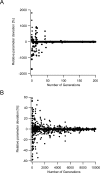
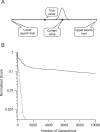
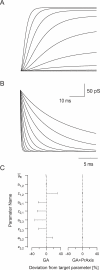
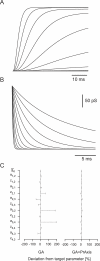

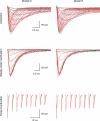
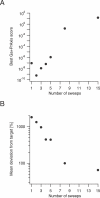


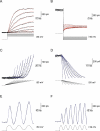
Similar articles
-
Mechanism for neuronal spike generation by small and large ion channel clusters.Phys Rev E Stat Nonlin Soft Matter Phys. 2004 Jul;70(1 Pt 1):011903. doi: 10.1103/PhysRevE.70.011903. Epub 2004 Jul 7. Phys Rev E Stat Nonlin Soft Matter Phys. 2004. PMID: 15324084
-
Evaluation of stochastic differential equation approximation of ion channel gating models.Ann Biomed Eng. 2009 Apr;37(4):824-38. doi: 10.1007/s10439-009-9635-z. Epub 2009 Jan 17. Ann Biomed Eng. 2009. PMID: 19152030
-
The use of automated parameter searches to improve ion channel kinetics for neural modeling.J Comput Neurosci. 2011 Oct;31(2):329-46. doi: 10.1007/s10827-010-0312-x. Epub 2011 Jan 18. J Comput Neurosci. 2011. PMID: 21243419
-
Channel noise in neurons.Trends Neurosci. 2000 Mar;23(3):131-7. doi: 10.1016/s0166-2236(99)01521-0. Trends Neurosci. 2000. PMID: 10675918 Review.
-
Molecular determinants of channel function.Physiol Rev. 1992 Oct;72(4 Suppl):S89-158. doi: 10.1152/physrev.1992.72.suppl_4.S89. Physiol Rev. 1992. PMID: 1279736 Review.
Cited by
-
A systems-biology approach to molecular machines: Exploration of alternative transporter mechanisms.PLoS Comput Biol. 2020 Jul 2;16(7):e1007884. doi: 10.1371/journal.pcbi.1007884. eCollection 2020 Jul. PLoS Comput Biol. 2020. PMID: 32614821 Free PMC article.
-
Inversion and computational maturation of drug response using human stem cell derived cardiomyocytes in microphysiological systems.Sci Rep. 2018 Dec 4;8(1):17626. doi: 10.1038/s41598-018-35858-7. Sci Rep. 2018. PMID: 30514966 Free PMC article.
-
Probing the dynamics of identified neurons with a data-driven modeling approach.PLoS One. 2008 Jul 9;3(7):e2627. doi: 10.1371/journal.pone.0002627. PLoS One. 2008. PMID: 18612435 Free PMC article.
-
Parameterization for In-Silico Modeling of Ion Channel Interactions with Drugs.PLoS One. 2016 Mar 10;11(3):e0150761. doi: 10.1371/journal.pone.0150761. eCollection 2016. PLoS One. 2016. PMID: 26963710 Free PMC article.
-
Cell-specific cardiac electrophysiology models.PLoS Comput Biol. 2015 Apr 30;11(4):e1004242. doi: 10.1371/journal.pcbi.1004242. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25928268 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

