Mining and state-space modeling and verification of sub-networks from large-scale biomolecular networks
- PMID: 17764552
- PMCID: PMC2213691
- DOI: 10.1186/1471-2105-8-324
Mining and state-space modeling and verification of sub-networks from large-scale biomolecular networks
Abstract
Background: Biomolecular networks dynamically respond to stimuli and implement cellular function. Understanding these dynamic changes is the key challenge for cell biologists. As biomolecular networks grow in size and complexity, the model of a biomolecular network must become more rigorous to keep track of all the components and their interactions. In general this presents the need for computer simulation to manipulate and understand the biomolecular network model.
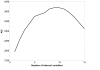
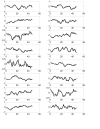
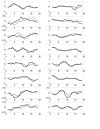
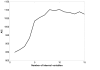
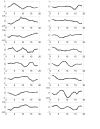
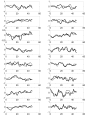
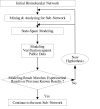
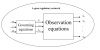
Results: In this paper, we present a novel method to model the regulatory system which executes a cellular function and can be represented as a biomolecular network. Our method consists of two steps. First, a novel scale-free network clustering approach is applied to the large-scale biomolecular network to obtain various sub-networks. Second, a state-space model is generated for the sub-networks and simulated to predict their behavior in the cellular context. The modeling results represent hypotheses that are tested against high-throughput data sets (microarrays and/or genetic screens) for both the natural system and perturbations. Notably, the dynamic modeling component of this method depends on the automated network structure generation of the first component and the sub-network clustering, which are both essential to make the solution tractable.
Conclusion: Experimental results on time series gene expression data for the human cell cycle indicate our approach is promising for sub-network mining and simulation from large-scale biomolecular network.
Figures


Similar articles
-
Mining, modeling, and evaluation of subnetworks from large biomolecular networks and its comparison study.IEEE Trans Inf Technol Biomed. 2009 Mar;13(2):184-94. doi: 10.1109/TITB.2008.2007649. IEEE Trans Inf Technol Biomed. 2009. PMID: 19272861
-
Identification of functional modules using network topology and high-throughput data.BMC Syst Biol. 2007 Jan 26;1:8. doi: 10.1186/1752-0509-1-8. BMC Syst Biol. 2007. PMID: 17408515 Free PMC article.
-
Network legos: building blocks of cellular wiring diagrams.J Comput Biol. 2008 Sep;15(7):829-44. doi: 10.1089/cmb.2007.0139. J Comput Biol. 2008. PMID: 18707557
-
Network integration and graph analysis in mammalian molecular systems biology.IET Syst Biol. 2008 Sep;2(5):206-21. doi: 10.1049/iet-syb:20070075. IET Syst Biol. 2008. PMID: 19045817 Free PMC article. Review.
-
Causal Queries from Observational Data in Biological Systems via Bayesian Networks: An Empirical Study in Small Networks.Methods Mol Biol. 2019;1883:111-142. doi: 10.1007/978-1-4939-8882-2_5. Methods Mol Biol. 2019. PMID: 30547398 Review.
Cited by
-
DeGNServer: deciphering genome-scale gene networks through high performance reverse engineering analysis.Biomed Res Int. 2013;2013:856325. doi: 10.1155/2013/856325. Epub 2013 Nov 17. Biomed Res Int. 2013. PMID: 24328032 Free PMC article.
References
-
- Newman MEJ. The structure and function of complex networks. SIAM Review. 2003;45:167–256.
-
- Hashimoto RF, Kim S, Shmulevich I, Zhang W, Bittner ML, Dougherty ER. Growing genetic regulatory networks from seed genes. Bioinformatics. 2004;20:1241–1247. - PubMed
-
- Flake GW, Lawrence SR, Giles CL, Coetzee FM. Self-organization and identification of web communities. IEEE Computer. 2002;35:66–71.
-
- Jansen R, Lan N, Qian J, Gerstein M. Integration of genomic datasets to predict protein complexes in yeast. J Struct Functional Genomics. 2002;2:71–81. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

