Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions
- PMID: 17762862
- PMCID: PMC2230676
- DOI: 10.1038/sj.emboj.7601841
Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions
Abstract
Hepatitis B virus (HBV) is a major human pathogen causing about 750,000 deaths per year. The virion consists of a nucleocapsid and an envelope formed by lipids, and three integral membrane proteins. Although we have detailed structural insights into the organization of the HBV core, the arrangement of the envelope in virions and its interaction with the nucleocapsid is elusive. Here we show the ultrastructure of hepatitis B virions purified from patient serum. We identified two morphological phenotypes, which appear as compact and gapped particles with nucleocapsids in distinguishable conformations. The overall structures of these nucleocapsids resemble recombinant cores with two alpha-helical spikes per asymmetric unit. At the charged tips the spikes are contacted by defined protrusions of the envelope proteins, probably via electrostatic interactions. The HBV envelope in the two morphotypes is to some extent variable, but the surface proteins follow a general packing scheme with up to three surface protein dimers per asymmetric unit. The variability in the structure of the envelope indicates that the nucleocapsid does not firmly constrain the arrangement of the surface proteins, but provides a general template for the packing.
Figures


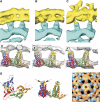
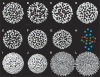
Similar articles
-
Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle.Virology. 1997 Feb 3;228(1):115-20. doi: 10.1006/viro.1996.8367. Virology. 1997. PMID: 9024817
-
Common and Distinct Capsid and Surface Protein Requirements for Secretion of Complete and Genome-Free Hepatitis B Virions.J Virol. 2018 Jun 29;92(14):e00272-18. doi: 10.1128/JVI.00272-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29743374 Free PMC article.
-
Analysis of the cytosolic domains of the hepatitis B virus envelope proteins for their function in viral particle assembly and infectivity.J Virol. 2006 Dec;80(24):11935-45. doi: 10.1128/JVI.00621-06. Epub 2006 Oct 4. J Virol. 2006. PMID: 17020942 Free PMC article.
-
Envelopment of the hepatitis B virus nucleocapsid.Virus Res. 2004 Dec;106(2):199-209. doi: 10.1016/j.virusres.2004.08.016. Virus Res. 2004. PMID: 15567498 Review.
-
[Envelope and core proteins of HBV and their role on the development of hepatitis B].Postepy Biochem. 1994;40(3):143-9. Postepy Biochem. 1994. PMID: 7937403 Review. Polish. No abstract available.
Cited by
-
Interaction of human tumor viruses with host cell surface receptors and cell entry.Viruses. 2015 May 22;7(5):2592-617. doi: 10.3390/v7052592. Viruses. 2015. PMID: 26008702 Free PMC article. Review.
-
Entry Inhibitors of Hepatitis B and D Viruses.Adv Exp Med Biol. 2022;1366:199-205. doi: 10.1007/978-981-16-8702-0_12. Adv Exp Med Biol. 2022. PMID: 35412142 Review.
-
Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein.Proc Natl Acad Sci U S A. 2013 Jul 23;110(30):E2782-91. doi: 10.1073/pnas.1308846110. Epub 2013 Jul 3. Proc Natl Acad Sci U S A. 2013. PMID: 23824290 Free PMC article.
-
Molecular Mechanisms to Control Post-Transplantation Hepatitis B Recurrence.Int J Mol Sci. 2015 Jul 30;16(8):17494-513. doi: 10.3390/ijms160817494. Int J Mol Sci. 2015. PMID: 26263973 Free PMC article. Review.
-
Direct interaction between the hepatitis B virus core and envelope proteins analyzed in a cellular context.Sci Rep. 2019 Nov 7;9(1):16178. doi: 10.1038/s41598-019-52824-z. Sci Rep. 2019. PMID: 31700077 Free PMC article.
References
-
- Böttcher B, Vogel M, Ploss M, Nassal M (2006) High plasticity of the hepatitis B virus capsid revealed by conformational stress. J Mol Biol 356: 812–822 - PubMed
-
- Böttcher B, Wynne SA, Crowther RA (1997) Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386: 88–91 - PubMed
-
- Bruss V (2004) Envelopment of the hepatitis B virus nucleocapsid. Virus Res 106: 199–209 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

