Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis
- PMID: 17726514
- PMCID: PMC1949146
- DOI: 10.1371/journal.pone.0000784
Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis
Abstract
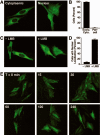
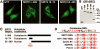
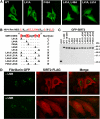
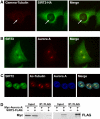
The human NAD+-dependent protein deacetylase SIRT2 resides predominantly in the cytoplasm where it functions as a tubulin deacetylase. Here we report that SIRT2 maintains a largely cytoplasmic localization during interphase by active nuclear export in a Crm1-dependent manner. We identified a functional, leptomycin B-sensitive, nuclear export signal sequence within SIRT2. During the cell cycle, SIRT2 becomes enriched in the nucleus and is associated with mitotic structures, beginning with the centrosome during prophase, the mitotic spindle during metaphase, and the midbody during cytokinesis. Cells overexpressing wild-type or a catalytically inactive SIRT2 exhibit an increase in multinucleated cells. The findings suggest a novel mechanism of regulating SIRT2 function by nucleo-cytoplasmic shuttling, as well as a role for SIRT2 in the nucleus during interphase and throughout mitosis.
Conflict of interest statement
Figures

Similar articles
-
Active nuclear import of the deacetylase Sirtuin-2 is controlled by its C-terminus and importins.Sci Rep. 2020 Feb 10;10(1):2034. doi: 10.1038/s41598-020-58397-6. Sci Rep. 2020. PMID: 32042025 Free PMC article.
-
SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress.Oncogene. 2007 Feb 15;26(7):945-57. doi: 10.1038/sj.onc.1209857. Epub 2006 Aug 14. Oncogene. 2007. PMID: 16909107
-
A putative N-terminal nuclear export sequence is sufficient for Mps1 nuclear exclusion during interphase.BMC Cell Biol. 2015 Feb 27;16:6. doi: 10.1186/s12860-015-0048-6. BMC Cell Biol. 2015. PMID: 25886724 Free PMC article.
-
Nucleoporins: leaving the nuclear pore complex for a successful mitosis.Cell Signal. 2011 Oct;23(10):1555-62. doi: 10.1016/j.cellsig.2011.05.023. Epub 2011 Jun 13. Cell Signal. 2011. PMID: 21683138 Review.
-
SIRT2: Controversy and multiple roles in disease and physiology.Ageing Res Rev. 2019 Nov;55:100961. doi: 10.1016/j.arr.2019.100961. Epub 2019 Sep 7. Ageing Res Rev. 2019. PMID: 31505260 Review.
Cited by
-
Host sirtuin 2 as an immunotherapeutic target against tuberculosis.Elife. 2020 Jul 22;9:e55415. doi: 10.7554/eLife.55415. Elife. 2020. PMID: 32697192 Free PMC article.
-
SIRT2 as a Therapeutic Target for Age-Related Disorders.Front Pharmacol. 2012 May 3;3:82. doi: 10.3389/fphar.2012.00082. eCollection 2012. Front Pharmacol. 2012. PMID: 22563317 Free PMC article.
-
Implications of altered sirtuins in metabolic regulation and oral cancer.PeerJ. 2023 Feb 14;11:e14752. doi: 10.7717/peerj.14752. eCollection 2023. PeerJ. 2023. PMID: 36815979 Free PMC article. Review.
-
Cysteine thiol oxidation on SIRT2 regulates inflammation in obese mice with sepsis.Inflammation. 2019 Feb;42(1):156-169. doi: 10.1007/s10753-018-0881-9. Inflammation. 2019. PMID: 30203196 Free PMC article.
-
Development of a NanoBRET assay to validate inhibitors of Sirt2-mediated lysine deacetylation and defatty-acylation that block prostate cancer cell migration.RSC Chem Biol. 2022 Mar 1;3(4):468-485. doi: 10.1039/d1cb00244a. eCollection 2022 Apr 6. RSC Chem Biol. 2022. PMID: 35441145 Free PMC article.
References
-
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. - PubMed
-
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. - PubMed
-
- Grubisha O, Smith BC, Denu JM. Small molecule regulation of Sir2 protein deacetylases. Febs J. 2005;272:4607–4616. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

