Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants
- PMID: 17715215
- PMCID: PMC2168797
- DOI: 10.1128/JVI.01525-07
Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants
Abstract
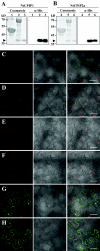
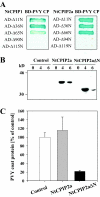
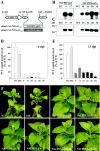
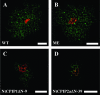
The capsid protein (CP) of potyviruses is required for various steps during plant infection, such as virion assembly, cell-to-cell movement, and long-distance transport. This suggests a series of compatible interactions with putative host factors which, however, are largely unknown. By using the yeast two-hybrid system the CP from Potato virus Y (PVY) was found to interact with a novel subset of DnaJ-like proteins from tobacco, designated NtCPIPs. Mutational analysis identified the CP core region, previously shown to be essential for virion formation and plasmodesmal trafficking, as the interacting domain. The ability of NtCPIP1 and NtCPIP2a to associate with PVY CP could be confirmed in vitro and was additionally verified in planta by bimolecular fluorescence complementation. The biological significance of the interaction was assayed by PVY infection of agroinfiltrated leaves and transgenic tobacco plants that expressed either full-length or J-domain-deficient variants of NtCPIPs. Transient expression of truncated dominant-interfering NtCPIP2a but not of the functional protein resulted in strongly reduced accumulation of PVY in the inoculated leaf. Consistently, stable overexpression of J-domain-deficient variants of NtCPIP1 and NtCPIP2a dramatically increased the virus resistance of various transgenic lines, indicating a critical role of functional NtCPIPs during PVY infection. The negative effect of impaired NtCPIP function on viral pathogenicity seemed to be the consequence of delayed cell-to-cell movement, as visualized by microprojectile bombardment with green fluorescent protein-tagged PVY. Therefore, we propose that NtCPIPs act as important susceptibility factors during PVY infection, possibly by recruiting heat shock protein 70 chaperones for viral assembly and/or cellular spread.
Figures


Similar articles
-
Complementation of the movement-deficient mutations in potato virus X: potyvirus coat protein mediates cell-to-cell trafficking of C-terminal truncation but not deletion mutant of potexvirus coat protein.Virology. 2000 Apr 25;270(1):31-42. doi: 10.1006/viro.2000.0246. Virology. 2000. PMID: 10772977
-
Nicotiana benthamiana protein, NbPCIP1, interacting with Potato virus X coat protein plays a role as susceptible factor for viral infection.Virology. 2009 Apr 10;386(2):257-69. doi: 10.1016/j.virol.2008.12.044. Epub 2009 Feb 11. Virology. 2009. PMID: 19215953
-
Transgenic plant virus resistance mediated by untranslatable sense RNAs: expression, regulation, and fate of nonessential RNAs.Plant Cell. 1994 Oct;6(10):1441-53. doi: 10.1105/tpc.6.10.1441. Plant Cell. 1994. PMID: 7994177 Free PMC article.
-
Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene Ny in potato.Mol Plant Microbe Interact. 2013 Mar;26(3):297-305. doi: 10.1094/MPMI-09-12-0219-R. Mol Plant Microbe Interact. 2013. PMID: 23113714
-
The HC-pro protein of potato virus Y interacts with NtMinD of tobacco.Mol Plant Microbe Interact. 2007 Dec;20(12):1505-11. doi: 10.1094/MPMI-20-12-1505. Mol Plant Microbe Interact. 2007. PMID: 17990958
Cited by
-
Two homologous host proteins interact with potato virus X RNAs and CPs and affect viral replication and movement.Sci Rep. 2016 Jun 29;6:28743. doi: 10.1038/srep28743. Sci Rep. 2016. PMID: 27353522 Free PMC article.
-
Fine mapping a quantitative trait locus underlying seedling resistance to gummy stem blight using a residual heterozygous lines-derived strategy in cucumber.Front Plant Sci. 2022 Sep 2;13:968811. doi: 10.3389/fpls.2022.968811. eCollection 2022. Front Plant Sci. 2022. PMID: 36119620 Free PMC article.
-
The Role of the Phylogenetically Conserved Cochaperone Protein Droj2/DNAJA3 in NF-κB Signaling.J Biol Chem. 2015 Sep 25;290(39):23816-25. doi: 10.1074/jbc.M115.664193. Epub 2015 Aug 5. J Biol Chem. 2015. PMID: 26245905 Free PMC article.
-
StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana.Viruses. 2022 Mar 10;14(3):574. doi: 10.3390/v14030574. Viruses. 2022. PMID: 35336981 Free PMC article.
-
P3 and NIa-Pro of Turnip Mosaic Virus Are Independent Elicitors of Superinfection Exclusion.Viruses. 2023 Jun 28;15(7):1459. doi: 10.3390/v15071459. Viruses. 2023. PMID: 37515147 Free PMC article.
References
-
- Agranovsky, A. A., V. P. Boyko, A. V. Karasev, E. V. Koonin, and V. V. Dolja. 1991. Putative 65 kDa protein of beet yellows closterovirus is a homologue of HSP70 heat shock proteins. J. Mol. Biol. 217:603-610. - PubMed
-
- Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241-263. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

