Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae
- PMID: 17705828
- PMCID: PMC2039749
- DOI: 10.1186/1471-2199-8-71
Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae
Abstract
Background: The nonsense-mediated mRNA decay (NMD) pathway promotes the rapid degradation of mRNAs containing premature termination codons (PTCs). In yeast Saccharomyces cerevisiae, the activity of the NMD pathway depends on the recognition of the PTC by the translational machinery. Translation termination factors eRF1 (Sup45) and eRF3 (Sup35) participate not only in the last step of protein synthesis but also in mRNA degradation and translation initiation via interaction with such proteins as Pab1, Upf1, Upf2 and Upf3.
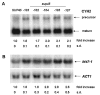
Results: In this work we have used previously isolated sup45 mutants of S. cerevisiae to characterize degradation of aberrant mRNA in conditions when translation termination is impaired. We have sequenced his7-1, lys9-A21 and trp1-289 alleles which are frequently used for analysis of nonsense suppression. We have established that sup45 nonsense and missense mutations lead to accumulation of his7-1 mRNA and CYH2 pre-mRNA. Remarkably, deletion of the UPF1 gene suppresses some sup45 phenotypes. In particular, sup45-n upf1Delta double mutants were less temperature sensitive, and more resistant to paromomycin than sup45 single mutants. In addition, deletion of either UPF2 or UPF3 restored viability of sup45-n double mutants.
Conclusion: This is the first demonstration that sup45 mutations do not only change translation fidelity but also acts by causing a change in mRNA stability.
Figures

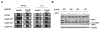


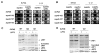
Similar articles
-
[Mutations in the Sup35 gene impairs degradation of mRNA containing premature stop codons].Mol Biol (Mosk). 2010 Jan-Feb;44(1):51-9. Mol Biol (Mosk). 2010. PMID: 20198859 Russian.
-
[The influence of UPF genes on the severity of SUP45 mutations].Mol Biol (Mosk). 2012 Mar-Apr;46(2):285-97. Mol Biol (Mosk). 2012. PMID: 22670525 Russian.
-
Role of RNA surveillance proteins Upf1/CpaR, Upf2 and Upf3 in the translational regulation of yeast CPA1 gene.Curr Genet. 2002 Jul;41(4):224-31. doi: 10.1007/s00294-002-0300-4. Epub 2002 Jun 15. Curr Genet. 2002. PMID: 12172963
-
UPF1 P-body localization.Biochem Soc Trans. 2008 Aug;36(Pt 4):698-700. doi: 10.1042/BST0360698. Biochem Soc Trans. 2008. PMID: 18631143 Review.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
Cited by
-
Regulation of release factor expression using a translational negative feedback loop: a systems analysis.RNA. 2012 Dec;18(12):2320-34. doi: 10.1261/rna.035113.112. Epub 2012 Oct 25. RNA. 2012. PMID: 23104998 Free PMC article.
-
Autoregulatory systems controlling translation factor expression: thermostat-like control of translational accuracy.RNA. 2010 Apr;16(4):655-63. doi: 10.1261/rna.1796210. Epub 2010 Feb 25. RNA. 2010. PMID: 20185543 Free PMC article. Review.
-
Mutation at tyrosine in AMLRY (GILRY like) motif of yeast eRF1 on nonsense codons suppression and binding affinity to eRF3.Int J Biol Sci. 2008 Apr 21;4(2):87-95. doi: 10.7150/ijbs.4.87. Int J Biol Sci. 2008. PMID: 18463713 Free PMC article.
-
The paradox of viable sup45 STOP mutations: a necessary equilibrium between translational readthrough, activity and stability of the protein.Mol Genet Genomics. 2009 Jul;282(1):83-96. doi: 10.1007/s00438-009-0447-5. Epub 2009 Apr 16. Mol Genet Genomics. 2009. PMID: 19370360
-
Modulation of efficiency of translation termination in Saccharomyces cerevisiae.Prion. 2014;8(3):247-60. doi: 10.4161/pri.29851. Epub 2014 Nov 1. Prion. 2014. PMID: 25486049 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

