Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation
- PMID: 17698638
- PMCID: PMC1986802
- DOI: 10.1261/rna.666807
Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation
Abstract
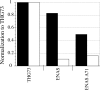
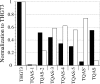
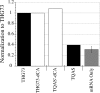
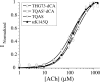
The incorporation of unnatural amino acids site-specifically is a valuable technique for structure-function studies, incorporation of biophysical probes, and determining protein-protein interactions. THG73 is an amber suppressor tRNA used extensively for the incorporation of >100 different residues in over 20 proteins, but under certain conditions THG73 is aminoacylated in vivo by endogenous aminoacyl-tRNA synthetase. Similar aminoacylation is seen with the Escherichia coli Asn amber suppressor tRNA, which has also been used to incorporate UAAs in many studies. We now find that the natural amino acid placed on THG73 is Gln. Since the E. coli GlnRS recognizes positions in the acceptor stem, we made several acceptor stem mutations in the second to fourth positions on THG73. All mutations reduce aminoacylation in vivo and allow for the selection of highly orthogonal tRNAs. To show the generality of these mutations, we created opal suppressor tRNAs that show less aminoacylation in Xenopus oocytes relative to THG73. We have created a library of Tetrahymena thermophila Gln amber suppressor tRNAs that will be useful for determining optimal suppressor tRNAs for use in other eukaryotic cells.
Figures
Similar articles
-
Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: evaluating suppression efficiency.RNA. 2007 Oct;13(10):1715-22. doi: 10.1261/rna.667607. Epub 2007 Aug 13. RNA. 2007. PMID: 17698637 Free PMC article.
-
In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression.Proc Natl Acad Sci U S A. 2006 Jun 6;103(23):8650-5. doi: 10.1073/pnas.0510817103. Epub 2006 May 25. Proc Natl Acad Sci U S A. 2006. PMID: 16728509 Free PMC article.
-
Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: concomitant suppression of three different termination codons in an mRNA in mammalian cells.Nucleic Acids Res. 2004 Dec 1;32(21):6200-11. doi: 10.1093/nar/gkh959. Print 2004. Nucleic Acids Res. 2004. PMID: 15576346 Free PMC article.
-
Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts.Acc Chem Res. 2017 Nov 21;50(11):2767-2775. doi: 10.1021/acs.accounts.7b00376. Epub 2017 Oct 6. Acc Chem Res. 2017. PMID: 28984438 Free PMC article. Review.
-
[The tRNA anticodon is recognized by aminoacyl-tRNA-synthetase].Mol Biol (Mosk). 1989 Nov-Dec;23(6):1603-10. Mol Biol (Mosk). 1989. PMID: 2698995 Review. Russian.
Cited by
-
Suppressor Mutants: History and Today's Applications.EcoSal Plus. 2021 Dec 15;9(2):eESP00372020. doi: 10.1128/ecosalplus.ESP-0037-2020. Epub 2021 Dec 15. EcoSal Plus. 2021. PMID: 34910591 Free PMC article. Review.
-
Chemical scale studies of the Phe-Pro conserved motif in the cys loop of Cys loop receptors.J Biol Chem. 2010 Mar 19;285(12):8976-84. doi: 10.1074/jbc.M109.060939. Epub 2010 Jan 12. J Biol Chem. 2010. PMID: 20068044 Free PMC article.
-
Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: evaluating suppression efficiency.RNA. 2007 Oct;13(10):1715-22. doi: 10.1261/rna.667607. Epub 2007 Aug 13. RNA. 2007. PMID: 17698637 Free PMC article.
-
An unusual pattern of ligand-receptor interactions for the α7 nicotinic acetylcholine receptor, with implications for the binding of varenicline.Mol Pharmacol. 2013 Aug;84(2):201-7. doi: 10.1124/mol.113.085795. Epub 2013 May 16. Mol Pharmacol. 2013. PMID: 23680636 Free PMC article.
-
The evolution of the ribosome and the genetic code.Life (Basel). 2014 May 20;4(2):227-49. doi: 10.3390/life4020227. Life (Basel). 2014. PMID: 25370196 Free PMC article.
References
-
- Anderson, J.C., Schultz, P.G. Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber, and opal suppression. Biochemistry. 2003;42:9598–9608. - PubMed
-
- Beene, D.L., Dougherty, D.A., Lester, H.A. Unnatural amino acid mutagenesis in mapping ion channel function. Curr. Opin. Neurobiol. 2003;13:264–270. - PubMed
-
- Brejc, K., van Dijk, W.J., Klaassen, R.V., Schuurmans, M., van Der Oost, J., Smit, A.B., Sixma, T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. - PubMed
-
- Capone, J.P., Sedivy, J.M., Sharp, P.A., RajBhandary, U.L. Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: Use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol. Cell. Biol. 1986;6:3059–3067. - PMC - PubMed
-
- Cload, S.T., Liu, D.R., Froland, W.A., Schultz, P.G. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem. Biol. 1996;3:1033–1038. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
