Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease
- PMID: 17696515
- PMCID: PMC2751596
- DOI: 10.1021/jm070482q
Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease
Abstract
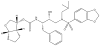
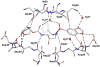
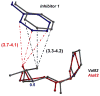
The potent new antiviral inhibitor GRL-98065 (1) of HIV-1 protease (PR) has been studied with PR variants containing the single mutations D30N, I50V, V82A, and I84V that provide resistance to the major clinical inhibitors. Compound 1 had inhibition constants of 17-fold, 8-fold, 3-fold, and 3-fold, respectively, for PR(D30N), PR(I50V), PR(V82A), and PR(I84V) relative to wild type PR. The chemically related darunavir had similar relative inhibition, except for PR(D30N), where inhibitor 1 was approximately 3-fold less potent. The high resolution (1.11-1.60 Angstrom) crystal structures of PR mutant complexes with inhibitor 1 showed small changes relative to the wild type enzyme. PR(D30N) and PR(V82A) showed compensating interactions with inhibitor 1 relative to those of PR, while reduced hydrophobic contacts were observed with PR(I50V) and PR(I84V). Importantly, inhibitor 1 complexes showed fewer changes relative to wild type enzyme than reported for darunavir complexes. Therefore, inhibitor 1 is a valuable addition to the antiviral inhibitors with high potency against resistant strains of HIV.
Figures








Similar articles
-
Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M.J Med Chem. 2006 Feb 23;49(4):1379-87. doi: 10.1021/jm050943c. J Med Chem. 2006. PMID: 16480273 Free PMC article.
-
High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains.J Mol Biol. 2004 Apr 23;338(2):341-52. doi: 10.1016/j.jmb.2004.02.052. J Mol Biol. 2004. PMID: 15066436
-
Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.FEBS J. 2005 Oct;272(20):5265-77. doi: 10.1111/j.1742-4658.2005.04923.x. FEBS J. 2005. PMID: 16218957 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
Cited by
-
Prediction of potency of protease inhibitors using free energy simulations with polarizable quantum mechanics-based ligand charges and a hybrid water model.J Chem Inf Model. 2009 Dec;49(12):2851-62. doi: 10.1021/ci900320p. J Chem Inf Model. 2009. PMID: 19928916 Free PMC article.
-
Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies.J Med Chem. 2008 Oct 9;51(19):6021-33. doi: 10.1021/jm8004543. Epub 2008 Sep 11. J Med Chem. 2008. PMID: 18783203 Free PMC article.
-
Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1'-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies.J Med Chem. 2009 Jul 9;52(13):3902-14. doi: 10.1021/jm900303m. J Med Chem. 2009. PMID: 19473017 Free PMC article.
-
Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS.J Med Chem. 2016 Jun 9;59(11):5172-208. doi: 10.1021/acs.jmedchem.5b01697. Epub 2016 Jan 22. J Med Chem. 2016. PMID: 26799988 Free PMC article. Review.
-
New approaches to HIV protease inhibitor drug design II: testing the substrate envelope hypothesis to avoid drug resistance and discover robust inhibitors.Curr Opin HIV AIDS. 2008 Nov;3(6):642-6. doi: 10.1097/COH.0b013e3283136cee. Curr Opin HIV AIDS. 2008. PMID: 19373036 Free PMC article.
References
-
- Barbaro G, Lucchini A, Barbarini G. Highly active antiretroviral therapy in HIV-associated pulmonary hypertension. Minerva Cardioangiol. 2005;53:153–4. - PubMed
-
- Barlett JA, DeMasi R, Quinn J, Moxham C, Rousseau F. Overview of the Effectiveness of Triple Combination Therapy in Antiretroviral-Naïve HIV-1 Infected Adults. AIDS. 2001;15:1369–1377. - PubMed
-
- Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–60. - PubMed
-
- Grabar S, Pradier C, Le Corfec E, Lancar R, Allavena C, Bentata M, Berlureau P, Dupont C, Fabbro-Peray P, Poizot-Martin I, Costagliola D. Factors associated with clinical and virological failure in patients receiving a triple therapy including a protease inhibitor. Aids. 2000;14:141–9. - PubMed
-
- Hertogs K, Bloor S, Kemp SD, Van den Eynde C, Alcorn TM, Pauwels R, Van Houtte M, Staszewski S, Miller V, Larder BA. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS. 2000;14:1203–1210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

