Antiviral immunity directed by small RNAs
- PMID: 17693253
- PMCID: PMC2703654
- DOI: 10.1016/j.cell.2007.07.039
Antiviral immunity directed by small RNAs
Abstract
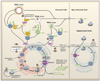
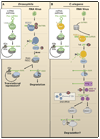
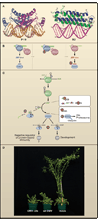
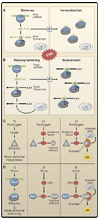
Plants and invertebrates can protect themselves from viral infection through RNA silencing. This antiviral immunity involves production of virus-derived small interfering RNAs (viRNAs) and results in specific silencing of viruses by viRNA-guided effector complexes. The proteins required for viRNA production as well as several key downstream components of the antiviral immunity pathway have been identified in plants, flies, and worms. Meanwhile, viral mechanisms to suppress this small RNA-directed immunity by viruses are being elucidated, thereby illuminating an ongoing molecular arms race that likely impacts the evolution of both viral and host genomes.
Figures
Similar articles
-
Antiviral innate immune response of RNA interference.J Infect Dev Ctries. 2014 Jul 14;8(7):804-10. doi: 10.3855/jidc.4187. J Infect Dev Ctries. 2014. PMID: 25022288 Review.
-
Virus counterdefense: diverse strategies for evading the RNA-silencing immunity.Annu Rev Microbiol. 2006;60:503-31. doi: 10.1146/annurev.micro.60.080805.142205. Annu Rev Microbiol. 2006. PMID: 16768647 Free PMC article. Review.
-
Induction and suppression of RNA silencing: insights from viral infections.Nat Rev Genet. 2005 Mar;6(3):206-20. doi: 10.1038/nrg1555. Nat Rev Genet. 2005. PMID: 15703763 Review.
-
RNA-based antiviral immunity.Nat Rev Immunol. 2010 Sep;10(9):632-44. doi: 10.1038/nri2824. Epub 2010 Aug 13. Nat Rev Immunol. 2010. PMID: 20706278 Review.
-
Plant RNA silencing in viral defence.Adv Exp Med Biol. 2011;722:39-58. doi: 10.1007/978-1-4614-0332-6_3. Adv Exp Med Biol. 2011. PMID: 21915781 Review.
Cited by
-
Characterization by Small RNA Sequencing of Taro Bacilliform CH Virus (TaBCHV), a Novel Badnavirus.PLoS One. 2015 Jul 24;10(7):e0134147. doi: 10.1371/journal.pone.0134147. eCollection 2015. PLoS One. 2015. PMID: 26207896 Free PMC article.
-
Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco.Planta. 2016 Oct;244(4):961-9. doi: 10.1007/s00425-016-2567-6. Epub 2016 Jul 25. Planta. 2016. PMID: 27456838
-
WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis.Proc Natl Acad Sci U S A. 2013 May 21;110(21):E1963-71. doi: 10.1073/pnas.1221347110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650359 Free PMC article.
-
Comparative analysis among the small RNA populations of source, sink and conductive tissues in two different plant-virus pathosystems.BMC Genomics. 2015 Feb 22;16(1):117. doi: 10.1186/s12864-015-1327-5. BMC Genomics. 2015. PMID: 25765188 Free PMC article.
-
Dynamics of defense-related components in two contrasting genotypes of tomato upon infection with Tomato Leaf Curl New Delhi Virus.Mol Biotechnol. 2012 Oct;52(2):140-50. doi: 10.1007/s12033-011-9481-8. Mol Biotechnol. 2012. PMID: 22161255
References
-
- Anandalakshmi R, Marathe R, Ge X, Herr JM, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bian XY, Rasheed MS, Seemanpillai MJ, Rezaian MA. Analysis of Silencing escape of Tomato leaf curl virus: an evaluation of the role of DNA methylation. Mol. Plant Microbe Interact. 2006;19:614–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

