Targeting of FAK Ser910 by ERK5 and PP1delta in non-stimulated and phorbol ester-stimulated cells
- PMID: 17692050
- PMCID: PMC2049076
- DOI: 10.1042/BJ20070058
Targeting of FAK Ser910 by ERK5 and PP1delta in non-stimulated and phorbol ester-stimulated cells
Abstract
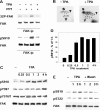
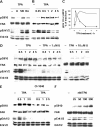
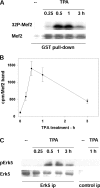
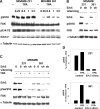
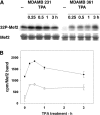
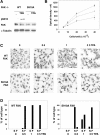
Ser910 of FAK (focal adhesion kinase) was phosphorylated in fibroblasts treated with the phorbol ester PMA and dephosphorylated by PP1d (protein phosphatase 1d), as indicated by shRNA (small-hairpin RNA) gene silencing. Ser910 of FAK was reported previously to be an ERK (extracellular-signal-regulated kinase) 1/2 target in cells treated with phorbol esters. In contrast, various approaches, including the use of the MEK (mitogen-activated protein kinase/ERK kinase) inhibitors UO126 and CI-1040 to inhibit ERK1/2 pointed to the involvement of ERK5. This hypothesis was confirmed by: (i) shRNA ERK5 gene silencing, which resulted in complete pSer910 loss in non-stimulated and PMA-stimulated cells; (ii) direct phosphorylation of recombinant FAK by ERK5; and (iii) ERK5 activation by PMA. PMA stimulation and ERK5 silencing in MDA-MB 231 and MDA-MB 361 breast cancer cells indicated Ser910 targeting by ERK5 also in these cells. Given the proximity of Ser910 to the FAT (focal adhesion targeting) regulatory domain of FAK, cell proliferation and morphology were investigated in FAK-/- cells expressing S910A mutant FAK. The cell growth rate decreased and exposure to PMA induced peculiar morphological changes in cells expressing S910A, with respect to wild-type FAK, suggesting a role for Ser910 in these processes. The present study indicates, for the first time, the phosphorylation of Ser910 of FAK by ERK5 and its dephosphorylation by PP1d, and suggested a role for Ser910 in the control of cell shape and proliferation.
Figures

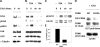
Similar articles
-
An altered fibronectin matrix induces anoikis of human squamous cell carcinoma cells by suppressing integrin alpha v levels and phosphorylation of FAK and ERK.Apoptosis. 2007 Dec;12(12):2221-31. doi: 10.1007/s10495-007-0138-9. Apoptosis. 2007. PMID: 17879163
-
Betagamma subunits of G(i/o) suppress EGF-induced ERK5 phosphorylation, whereas ERK1/2 phosphorylation is enhanced.Cell Signal. 2008 Jul;20(7):1275-83. doi: 10.1016/j.cellsig.2008.02.016. Epub 2008 Mar 4. Cell Signal. 2008. PMID: 18407464
-
Basic fibroblast growth factor promotes glial cell-derived neurotrophic factor gene expression mediated by activation of ERK5 in rat C6 glioma cells.Cell Signal. 2011 Apr;23(4):666-72. doi: 10.1016/j.cellsig.2010.11.020. Epub 2010 Dec 2. Cell Signal. 2011. PMID: 21130871
-
FAK expression regulation and therapeutic potential.Adv Cancer Res. 2008;101:45-61. doi: 10.1016/S0065-230X(08)00403-X. Adv Cancer Res. 2008. PMID: 19055942 Review.
-
Signal transduction by focal adhesion kinase in cancer.Cancer Metastasis Rev. 2009 Jun;28(1-2):35-49. doi: 10.1007/s10555-008-9165-4. Cancer Metastasis Rev. 2009. PMID: 19169797 Review.
Cited by
-
Clinical Significance and Regulation of ERK5 Expression and Function in Cancer.Cancers (Basel). 2022 Jan 11;14(2):348. doi: 10.3390/cancers14020348. Cancers (Basel). 2022. PMID: 35053510 Free PMC article. Review.
-
ERK5 suppression overcomes FAK inhibitor resistance in mutant KRAS-driven non-small cell lung cancer.EMBO Mol Med. 2024 Oct;16(10):2402-2426. doi: 10.1038/s44321-024-00138-7. Epub 2024 Sep 13. EMBO Mol Med. 2024. PMID: 39271958 Free PMC article.
-
Oncogenic signaling of MEK5-ERK5.Cancer Lett. 2017 Apr 28;392:51-59. doi: 10.1016/j.canlet.2017.01.034. Epub 2017 Jan 30. Cancer Lett. 2017. PMID: 28153789 Free PMC article. Review.
-
FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST.Mol Cell. 2009 Jul 10;35(1):11-25. doi: 10.1016/j.molcel.2009.06.013. Mol Cell. 2009. PMID: 19595712 Free PMC article.
-
Serine-910 phosphorylation of focal adhesion kinase is critical for sarcomere reorganization in cardiomyocyte hypertrophy.Cardiovasc Res. 2011 Dec 1;92(3):409-19. doi: 10.1093/cvr/cvr247. Epub 2011 Sep 21. Cardiovasc Res. 2011. PMID: 21937583 Free PMC article.
References
-
- Hanks S. K., Ryzhova L., Shin N., Brabek J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front. Biosci. 2003;8:d982–d986. - PubMed
-
- Parsons J. T. Focal adhesion kinase: the first ten years. J. Cell Sci. 2003;116:1409–1416. - PubMed
-
- Carragher N. O., Frame M. C. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 2004;14:241–249. - PubMed
-
- Owen J. D., Ruest P. J., Fry D. W., Hanks S. K. Induced focal adhesion kinase (FAK) expression in FAK-null cells enhances cell spreading and migration requiring both auto- and activation loop phosphorylation sites and inhibits adhesion-dependent tyrosine phosphorylation of Pyk2. Mol. Cell. Biol. 1999;19:4806–4818. - PMC - PubMed
-
- Schlaepfer D. D., Mitra S. K., Ilic D. Control of motile and invasive phenotypes by focal adhesion kinase. Biochim. Biophys. Acta. 2004;1692:77–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

