Protégé: a tool for managing and using terminology in radiology applications
- PMID: 17687607
- PMCID: PMC2039856
- DOI: 10.1007/s10278-007-9065-0
Protégé: a tool for managing and using terminology in radiology applications
Abstract
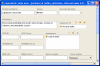
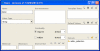
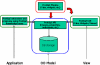
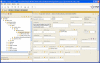
The development of standard terminologies such as RadLex is becoming important in radiology applications, such as structured reporting, teaching file authoring, report indexing, and text mining. The development and maintenance of these terminologies are challenging, however, because there are few specialized tools to help developers to browse, visualize, and edit large taxonomies. Protégé ( http://protege.stanford.edu ) is an open-source tool that allows developers to create and to manage terminologies and ontologies. It is more than a terminology-editing tool, as it also provides a platform for developers to use the terminologies in end-user applications. There are more than 70,000 registered users of Protégé who are using the system to manage terminologies and ontologies in many different domains. The RadLex project has recently adopted Protégé for managing its radiology terminology. Protégé provides several features particularly useful to managing radiology terminologies: an intuitive graphical user interface for navigating large taxonomies, visualization components for viewing complex term relationships, and a programming interface so developers can create terminology-driven radiology applications. In addition, Protégé has an extensible plug-in architecture, and its large user community has contributed a rich library of components and extensions that provide much additional useful functionalities. In this report, we describe Protégé's features and its particular advantages in the radiology domain in the creation, maintenance, and use of radiology terminology.
Figures



Similar articles
-
Creating and curating a terminology for radiology: ontology modeling and analysis.J Digit Imaging. 2008 Dec;21(4):355-62. doi: 10.1007/s10278-007-9073-0. Epub 2007 Sep 15. J Digit Imaging. 2008. PMID: 17874267 Free PMC article.
-
Protégé-2000: an open-source ontology-development and knowledge-acquisition environment.AMIA Annu Symp Proc. 2003;2003:953. AMIA Annu Symp Proc. 2003. PMID: 14728458 Free PMC article.
-
Design and implementation of an open source indexing solution for a large set of radiological reports and images.J Digit Imaging. 2007 Nov;20 Suppl 1(Suppl 1):11-20. doi: 10.1007/s10278-007-9055-2. Epub 2007 Aug 9. J Digit Imaging. 2007. PMID: 17687608 Free PMC article.
-
RadMonitor: radiology operations data mining in real time.J Digit Imaging. 2008 Sep;21(3):257-68. doi: 10.1007/s10278-007-9033-8. Epub 2007 May 30. J Digit Imaging. 2008. PMID: 17534683 Free PMC article. Review.
-
SimpleDICOM suite: personal productivity tools for managing DICOM objects.Radiographics. 2007 Sep-Oct;27(5):1523-30. doi: 10.1148/rg.275065139. Radiographics. 2007. PMID: 17848708 Review.
Cited by
-
Methods and applications for visualization of SNOMED CT concept sets.Appl Clin Inform. 2014 Feb 19;5(1):127-52. doi: 10.4338/ACI-2013-09-RA-0071. eCollection 2014. Appl Clin Inform. 2014. PMID: 24734129 Free PMC article.
-
KA-SB: from data integration to large scale reasoning.BMC Bioinformatics. 2009 Oct 1;10 Suppl 10(Suppl 10):S5. doi: 10.1186/1471-2105-10-S10-S5. BMC Bioinformatics. 2009. PMID: 19796402 Free PMC article.
-
Tools and collaborative environments for bioinformatics research.Brief Bioinform. 2011 Nov;12(6):549-61. doi: 10.1093/bib/bbr055. Epub 2011 Oct 7. Brief Bioinform. 2011. PMID: 21984743 Free PMC article.
-
PIBAS FedSPARQL: a web-based platform for integration and exploration of bioinformatics datasets.J Biomed Semantics. 2017 Sep 20;8(1):42. doi: 10.1186/s13326-017-0151-z. J Biomed Semantics. 2017. PMID: 28931422 Free PMC article.
-
Differential Adverse Event Profiles Associated with BCG as a Preventive Tuberculosis Vaccine or Therapeutic Bladder Cancer Vaccine Identified by Comparative Ontology-Based VAERS and Literature Meta-Analysis.PLoS One. 2016 Oct 17;11(10):e0164792. doi: 10.1371/journal.pone.0164792. eCollection 2016. PLoS One. 2016. PMID: 27749923 Free PMC article.
References
-
- Keizer NF, Abu-Hanna A, Cornet R, Zwetsloot-Schonk JH, Stoutenbeek CP. Analysis and design of an ontology for intensive care diagnoses. Methods Inf Med. 1999;38:102–112. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

