Transcriptional and epigenetic regulation of the integrin collagen receptor locus ITGA1-PELO-ITGA2
- PMID: 17669516
- PMCID: PMC2682338
- DOI: 10.1016/j.bbaexp.2007.06.004
Transcriptional and epigenetic regulation of the integrin collagen receptor locus ITGA1-PELO-ITGA2
Abstract
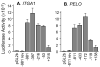
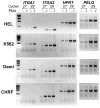
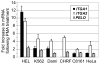
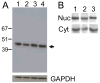
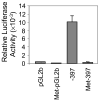
The integrin collagen receptor locus on human chromosome 5q11.2 includes the integrin genes ITGA1 and ITGA2, and the cell cycle regulation gene PELO, embedded within ITGA1 intron 1. ITGA1 contains a CArG box that is bound by serum response factor (SRF), while PELO contains two Sp1 binding elements. A comparison of mRNA levels in megakaryocytic (MK) and non-megakaryocytic (non-MK) cell lines and an analysis of the transcriptional activity of promoter-LUC reporter gene constructs in transfected cells revealed that ITGA1 is selectively suppressed in the MK lineage. Sodium bisulfite genomic sequencing established that a CpG-rich ITGA1 promoter region (-209/+115) is fully methylated at 19 CpG sites in MK cells that do not express alpha1beta1, but completely demethylated in expressing cells. In vitro methylation of ITGA1 suppresses transcription, while treatment of megakaryocytic cells with 5-aza-2'-deoxycytidine, but not Trichostatin A, resulted in de novo expression of ITGA1. During thrombopoietin-induced in vitro differentiation of primary human cord blood mononuclear cells into megakaryocytes, we observed rapid, progressive CpG methylation of ITGA1, but not PELO or ITGA2. Thus, selective CpG methylation of the ITGA1 promoter is a specific feature of alpha1beta1 regulation that coincides with the initiation of megakaryocyte differentiation.
Figures







Similar articles
-
Integrin α1β1 expression is controlled by c-MYC in colorectal cancer cells.Oncogene. 2016 Mar 31;35(13):1671-8. doi: 10.1038/onc.2015.231. Epub 2015 Jun 22. Oncogene. 2016. PMID: 26096932 Free PMC article.
-
Thrombopoietin initiates demethylation-based transcription of GP6 during megakaryocyte differentiation.Blood. 2005 May 15;105(10):3888-92. doi: 10.1182/blood-2004-08-3109. Epub 2005 Feb 8. Blood. 2005. PMID: 15701720 Free PMC article.
-
Epigenetic modifications affect Dnmt3L expression.Biochem J. 2004 Jun 15;380(Pt 3):705-13. doi: 10.1042/BJ20040067. Biochem J. 2004. PMID: 15015937 Free PMC article.
-
Binding of phosphorylated Sp1 protein to tandem Sp1 binding sites regulates alpha2 integrin gene core promoter activity.Blood. 1997 Jul 15;90(2):678-89. Blood. 1997. PMID: 9226168
-
Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression.J Clin Invest. 2004 Oct;114(8):1146-57. doi: 10.1172/JCI21647. J Clin Invest. 2004. PMID: 15489962 Free PMC article.
Cited by
-
PSAT1 Promotes Metastasis via p-AKT/SP1/ITGA2 Axis in Estrogen Receptor-Negative Breast Cancer Cell.Biomolecules. 2024 Aug 12;14(8):990. doi: 10.3390/biom14080990. Biomolecules. 2024. PMID: 39199378 Free PMC article.
-
Designating eukaryotic orthology via processed transcription units.Nucleic Acids Res. 2008 Jun;36(10):3436-42. doi: 10.1093/nar/gkn227. Epub 2008 Apr 29. Nucleic Acids Res. 2008. PMID: 18445630 Free PMC article.
-
Genome-wide association study of anthropometric traits in Korcula Island, Croatia.Croat Med J. 2009 Feb;50(1):7-16. doi: 10.3325/cmj.2009.50.7. Croat Med J. 2009. PMID: 19260139 Free PMC article.
-
Enhanced binding of poly(ADP-ribose)polymerase-1 and Ku80/70 to the ITGA2 promoter via an extended cytosine-adenosine repeat.PLoS One. 2010 Jan 15;5(1):e8743. doi: 10.1371/journal.pone.0008743. PLoS One. 2010. PMID: 20090957 Free PMC article.
-
RNA-seq transcriptome profiling of porcine lung from two pig breeds in response to Mycoplasma hyopneumoniae infection.PeerJ. 2019 Oct 21;7:e7900. doi: 10.7717/peerj.7900. eCollection 2019. PeerJ. 2019. PMID: 31656701 Free PMC article.
References
-
- Hughes AL. Evolution of the Integrin alpha and beta Protein Families. J Mol Evol. 2001;52:63. - PubMed
-
- Obata H, Hayashi K, Nishida W, Momiyama T, Uchida A, Ochi T, Sobue K. Smooth muscle cell phenotype-dependent transcriptional regulation of the alpha1 integrin gene. J Biol Chem. 1997;272:26643. - PubMed
-
- Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308. - PubMed
-
- Eberhart CG, Wasserman SA. The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development. 1995;121:3477. - PubMed
-
- Shamsadin R, Adham IM, Engel W. Mouse pelota gene (Pelo): cDNA cloning, genomic structure, and chromosomal localization. Cytogenet Genome Res. 2002;97:95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

