PHIDIAS: a pathogen-host interaction data integration and analysis system
- PMID: 17663773
- PMCID: PMC2323235
- DOI: 10.1186/gb-2007-8-7-r150
PHIDIAS: a pathogen-host interaction data integration and analysis system
Abstract
The Pathogen-Host Interaction Data Integration and Analysis System (PHIDIAS) is a web-based database system that serves as a centralized source to search, compare, and analyze integrated genome sequences, conserved domains, and gene expression data related to pathogen-host interactions (PHIs) for pathogen species designated as high priority agents for public health and biological security. In addition, PHIDIAS allows submission, search and analysis of PHI genes and molecular networks curated from peer-reviewed literature. PHIDIAS is publicly available at http://www.phidias.us.
Figures
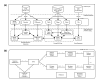






Similar articles
-
EHFPI: a database and analysis resource of essential host factors for pathogenic infection.Nucleic Acids Res. 2015 Jan;43(Database issue):D946-55. doi: 10.1093/nar/gku1086. Epub 2014 Nov 20. Nucleic Acids Res. 2015. PMID: 25414353 Free PMC article.
-
PHI-base: a new interface and further additions for the multi-species pathogen-host interactions database.Nucleic Acids Res. 2017 Jan 4;45(D1):D604-D610. doi: 10.1093/nar/gkw1089. Epub 2016 Dec 3. Nucleic Acids Res. 2017. PMID: 27915230 Free PMC article.
-
PHI-base: the pathogen-host interactions database.Nucleic Acids Res. 2020 Jan 8;48(D1):D613-D620. doi: 10.1093/nar/gkz904. Nucleic Acids Res. 2020. PMID: 31733065 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Progress in computational studies of host-pathogen interactions.J Bioinform Comput Biol. 2013 Apr;11(2):1230001. doi: 10.1142/S0219720012300018. Epub 2012 Oct 24. J Bioinform Comput Biol. 2013. PMID: 23600809 Review.
Cited by
-
Methanobacterium formicicum as a target rumen methanogen for the development of new methane mitigation interventions: A review.Vet Anim Sci. 2018 Sep 13;6:86-94. doi: 10.1016/j.vas.2018.09.001. eCollection 2018 Dec. Vet Anim Sci. 2018. PMID: 32734058 Free PMC article. Review.
-
Comparative genomics analysis to differentiate metabolic and virulence gene potential in gastric versus enterohepatic Helicobacter species.BMC Genomics. 2018 Nov 20;19(1):830. doi: 10.1186/s12864-018-5171-2. BMC Genomics. 2018. PMID: 30458713 Free PMC article.
-
Literature-based discovery of IFN-gamma and vaccine-mediated gene interaction networks.J Biomed Biotechnol. 2010;2010:426479. doi: 10.1155/2010/426479. Epub 2010 Jun 3. J Biomed Biotechnol. 2010. PMID: 20625487 Free PMC article.
-
Identification and functional analysis of AG1-IA specific genes of Rhizoctonia solani.Curr Genet. 2014 Nov;60(4):327-41. doi: 10.1007/s00294-014-0438-x. Epub 2014 Jul 29. Curr Genet. 2014. PMID: 25070039
-
A review on host-pathogen interactions: classification and prediction.Eur J Clin Microbiol Infect Dis. 2016 Oct;35(10):1581-99. doi: 10.1007/s10096-016-2716-7. Epub 2016 Jul 29. Eur J Clin Microbiol Infect Dis. 2016. PMID: 27470504 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

