Evidence that RNA silencing functions as an antiviral defense mechanism in fungi
- PMID: 17646660
- PMCID: PMC1937564
- DOI: 10.1073/pnas.0702500104
Evidence that RNA silencing functions as an antiviral defense mechanism in fungi
Abstract
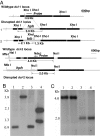
The role of RNA silencing as an antiviral defense mechanism in fungi was examined by testing the effect of dicer gene disruptions on mycovirus infection of the chestnut blight fungus Cryphonectria parasitica. C. parasitica dicer-like genes dcl-1 and dcl-2 were cloned and shown to share a high level of predicted amino acid sequence identity with the corresponding dicer-like genes from Neurospora crassa [Ncdcl-1 (50.5%); Ncdcl-2 (38.0%)] and Magnaporthe oryzae [MDL-1 (45.6%); MDL-2 (38.0%)], respectively. Disruption of dcl-1 and dcl-2 resulted in no observable phenotypic changes relative to wild-type C. parasitica. Infection of Deltadcl-1 strains with hypovirus CHV1-EP713 or reovirus MyRV1-Cp9B21 resulted in phenotypic changes that were indistinguishable from that exhibited by wild-type strain C. parasitica EP155 infected with these same viruses. In stark contrast, the Deltadcl-2 and Deltadcl-1/Deltadcl-2 mutant strains were highly susceptible to mycovirus infection, with CHV1-EP713-infected mutant strains becoming severely debilitated. Increased viral RNA levels were observed in the Deltadcl-2 mutant strains for a hypovirus CHV1-EP713 mutant lacking the suppressor of RNA silencing p29 and for wild-type reovirus MyRV1-Cp9B21. Complementation of the Deltadcl-2 strain with the wild-type dcl-2 gene resulted in reversion to the wild-type response to virus infection. These results provide direct evidence that a fungal dicer-like gene functions to regulate virus infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
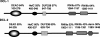


Similar articles
-
Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut blight fungus Cryphonectria parasitica.Eukaryot Cell. 2007 Aug;6(8):1286-98. doi: 10.1128/EC.00166-07. Epub 2007 Jun 8. Eukaryot Cell. 2007. PMID: 17557883 Free PMC article.
-
Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713.J Gen Virol. 2006 Dec;87(Pt 12):3703-3714. doi: 10.1099/vir.0.82213-0. J Gen Virol. 2006. PMID: 17098988
-
Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system.Eukaryot Cell. 2006 Jun;5(6):896-904. doi: 10.1128/EC.00373-05. Eukaryot Cell. 2006. PMID: 16757737 Free PMC article.
-
The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host.Virology. 2015 Mar;477:164-175. doi: 10.1016/j.virol.2014.09.024. Epub 2014 Nov 4. Virology. 2015. PMID: 25454384 Review.
-
[Cryphonectria parasitica as a host of fungal viruses: a tool useful to unravel the mycovirus world].Uirusu. 2014;64(1):11-24. doi: 10.2222/jsv.64.11. Uirusu. 2014. PMID: 25765976 Review. Japanese.
Cited by
-
Arthropod-borne flaviviruses and RNA interference: seeking new approaches for antiviral therapy.Adv Virus Res. 2013;85:91-111. doi: 10.1016/B978-0-12-408116-1.00004-5. Adv Virus Res. 2013. PMID: 23439025 Free PMC article. Review.
-
Diverse Partitiviruses From the Phytopathogenic Fungus, Rosellinia necatrix.Front Microbiol. 2020 Jun 26;11:1064. doi: 10.3389/fmicb.2020.01064. eCollection 2020. Front Microbiol. 2020. PMID: 32670213 Free PMC article.
-
Diverse, Novel Mycoviruses From the Virome of a Hypovirulent Sclerotium rolfsii Strain.Front Plant Sci. 2018 Nov 27;9:1738. doi: 10.3389/fpls.2018.01738. eCollection 2018. Front Plant Sci. 2018. PMID: 30542362 Free PMC article.
-
Discovery of microRNA-like RNAs during early fruiting body development in the model mushroom Coprinopsis cinerea.PLoS One. 2018 Sep 19;13(9):e0198234. doi: 10.1371/journal.pone.0198234. eCollection 2018. PLoS One. 2018. PMID: 30231028 Free PMC article.
-
A Satellite dsRNA Attenuates the Induction of Helper Virus-Mediated Symptoms in Aspergillus flavus.Front Microbiol. 2022 May 31;13:895844. doi: 10.3389/fmicb.2022.895844. eCollection 2022. Front Microbiol. 2022. PMID: 35711767 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

