Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation
- PMID: 17646391
- PMCID: PMC2099228
- DOI: 10.1128/MCB.01695-06
Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation
Abstract
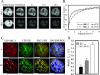
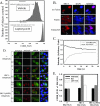
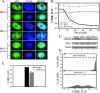
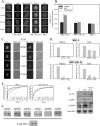
The steroid receptor coactivator 3 gene (SRC-3) (AIB1/ACTR/pCIP/RAC3/TRAM1) is a p160 family transcription coactivator and a known oncogene. Despite its importance, the functional regulation of SRC-3 remains poorly understood within a cellular context. Using a novel combination of live-cell, high-throughput, and fluorescent microscopy, we report SRC-3 to be a nucleocytoplasmic shuttling protein whose intracellular mobility, solubility, and cellular localization are regulated by phosphorylation and estrogen receptor alpha (ERalpha) interactions. We show that both chemical inhibition and small interfering RNA reduction of the mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 (MEK1/2) pathway induce a cytoplasmic shift in SRC-3 localization, whereas stimulation by epidermal growth factor signaling enhances its nuclear localization by inducing phosphorylation at T24, S857, and S860, known participants in the phosphocode that regulates SRC-3 activity. Accordingly, the cytoplasmic localization of a nonphosphorylatable SRC-3 mutant further supported these results. In the presence of ERalpha, U0126 also dramatically reduces (i) ligand-dependent colocalization of SRC-3 and ERalpha, (ii) the formation of ER-SRC-3 complexes in cell lysates, and (iii) SRC-3 targeting to a visible, ERalpha-occupied and -regulated prolactin promoter array. Taken together, these results indicate that phosphorylation coordinates SRC-3 coactivator function by linking the probabilistic formation of transient nuclear receptor-coactivator complexes with its molecular dynamics and cellular compartmentalization. Technically and conceptually, these findings have a new and broad impact upon evaluating mechanisms of action of gene regulators at a cellular system level.
Figures


Similar articles
-
Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-alpha: regulation by phosphorylation sites in the A/B region depends on other receptor domains.Mol Endocrinol. 2003 Jul;17(7):1296-314. doi: 10.1210/me.2001-0316. Epub 2003 Apr 24. Mol Endocrinol. 2003. PMID: 12714702
-
Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor.Mol Cell Biol. 2005 Sep;25(18):8273-84. doi: 10.1128/MCB.25.18.8273-8284.2005. Mol Cell Biol. 2005. PMID: 16135815 Free PMC article.
-
Isoform-selective interactions between estrogen receptors and steroid receptor coactivators promoted by estradiol and ErbB-2 signaling in living cells.Mol Endocrinol. 2003 Apr;17(4):589-99. doi: 10.1210/me.2002-0351. Epub 2003 Jan 16. Mol Endocrinol. 2003. PMID: 12554772
-
SRC-3/AIB1: transcriptional coactivator in oncogenesis.Acta Pharmacol Sin. 2006 Apr;27(4):387-94. doi: 10.1111/j.1745-7254.2006.00315.x. Acta Pharmacol Sin. 2006. PMID: 16539836 Review.
-
Review of the in vivo functions of the p160 steroid receptor coactivator family.Mol Endocrinol. 2003 Sep;17(9):1681-92. doi: 10.1210/me.2003-0116. Epub 2003 Jun 12. Mol Endocrinol. 2003. PMID: 12805412 Review.
Cited by
-
Homeostatic levels of SRC-2 and SRC-3 promote early human adipogenesis.J Cell Biol. 2011 Jan 10;192(1):55-67. doi: 10.1083/jcb.201004026. J Cell Biol. 2011. PMID: 21220509 Free PMC article.
-
SRC-3 has a role in cancer other than as a nuclear receptor coactivator.Int J Biol Sci. 2011;7(5):664-72. doi: 10.7150/ijbs.7.664. Epub 2011 May 24. Int J Biol Sci. 2011. PMID: 21647249 Free PMC article. Review.
-
Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells.Mol Cell Biol. 2008 Nov;28(21):6580-93. doi: 10.1128/MCB.00118-08. Epub 2008 Sep 2. Mol Cell Biol. 2008. PMID: 18765637 Free PMC article.
-
High content imaging-based assay to classify estrogen receptor-α ligands based on defined mechanistic outcomes.Gene. 2011 May 15;477(1-2):42-52. doi: 10.1016/j.gene.2011.01.009. Epub 2011 Jan 20. Gene. 2011. PMID: 21256200 Free PMC article.
-
Nuclear receptor coactivators: structural and functional biochemistry.Biochemistry. 2011 Jan 25;50(3):313-28. doi: 10.1021/bi101762x. Epub 2010 Dec 29. Biochemistry. 2011. PMID: 21141906 Free PMC article. Review.
References
-
- Amazit, L., Y. Alj, R. K. Tyagi, A. Chauchereau, H. Loosfelt, C. Pichon, J. Pantel, E. Foulon-Guinchard, P. Leclerc, E. Milgrom, and A. Guiochon-Mantel. 2003. Subcellular localization and mechanisms of nucleocytoplasmic trafficking of steroid receptor coactivator-1. J. Biol. Chem. 278:32195-32203. - PubMed
-
- Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. - PubMed
-
- Arimura, A., M. van Peer, A. J. Schroder, and P. B. Rothman. 2004. The transcriptional co-activator p/CIP (NCoA-3) is up-regulated by STAT6 and serves as a positive regulator of transcriptional activation by STAT6. J. Biol. Chem. 279:31105-31112. - PubMed
-
- Avivar, A., M. C. Garcia-Macias, E. Ascaso, G. Herrera, J. E. O'Connor, and J. F. de Mora. 2006. Moderate overexpression of AIB1 triggers pre-neoplastic changes in mammary epithelium. FEBS Lett. 580:5222-5226. - PubMed
-
- Baumann, C. T., H. Ma, R. Wolford, J. C. Reyes, P. Maruvada, C. Lim, P. M. Yen, M. R. Stallcup, and G. L. Hager. 2001. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485-500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
