Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis
- PMID: 17626615
- PMCID: PMC1941738
- DOI: 10.1186/1471-213X-7-84
Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis
Abstract
Background: Alternative promoters usage is an important paradigm in transcriptional control of mammalian gene expression. However, despite the growing interest in alternative promoters and their role in genome diversification, very little is known about how and on what occasions those promoters are differentially regulated. Runx1 transcription factor is a key regulator of early hematopoiesis and a frequent target of chromosomal translocations in acute leukemias. Mice deficient in Runx1 lack definitive hematopoiesis and die in mid-gestation. Expression of Runx1 is regulated by two functionally distinct promoters designated P1 and P2. Differential usage of these two promoters creates diversity in distribution and protein-coding potential of the mRNA transcripts. While the alternative usage of P1 and P2 likely plays an important role in Runx1 biology, very little is known about the function of the P1/P2 switch in mediating tissue and stage specific expression of Runx1 during development.
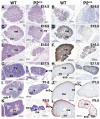
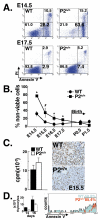
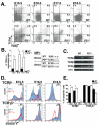
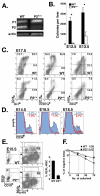
Results: We employed mice bearing a hypomorphic Runx1 allele, with a largely diminished P2 activity, to investigate the biological role of alternative P1/P2 usage. Mice homozygous for the hypomorphic allele developed to term, but died within a few days after birth. During embryogenesis the P1/P2 activity is spatially and temporally modulated. P2 activity is required in early hematopoiesis and when attenuated, development of liver hematopoietic progenitor cells (HPC) was impaired. Early thymus development and thymopoiesis were also abrogated as reflected by thymic hypocellularity and loss of corticomedullary demarcation. Differentiation of CD4/CD8 thymocytes was impaired and their apoptosis was enhanced due to altered expression of T-cell receptors.
Conclusion: The data delineate the activity of P1 and P2 in embryogenesis and describe previously unknown functions of Runx1. The findings show unequivocally that the role of P1/P2 during development is non redundant and underscore the significance of alternative promoter usage in Runx1 biology.
Figures



Similar articles
-
Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development.Biochem Biophys Res Commun. 2001 Mar;281(5):1248-55. doi: 10.1006/bbrc.2001.4513. Biochem Biophys Res Commun. 2001. PMID: 11243869
-
Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis.Blood. 2010 Apr 15;115(15):3042-50. doi: 10.1182/blood-2009-08-238626. Epub 2010 Feb 4. Blood. 2010. PMID: 20139099
-
Increase of hematopoietic progenitor and suppression of endothelial gene expression by Runx1 expression during in vitro ES differentiation.Exp Hematol. 2009 Mar;37(3):334-45. doi: 10.1016/j.exphem.2008.11.007. Exp Hematol. 2009. PMID: 19218012
-
Pathways in blood and vessel development revealed through zebrafish genetics.Int J Dev Biol. 2002;46(4):493-502. Int J Dev Biol. 2002. PMID: 12141436 Review.
-
The role of Runx1/AML1 and Evi-1 in the regulation of hematopoietic stem cells.J Cell Physiol. 2010 Feb;222(2):282-5. doi: 10.1002/jcp.21953. J Cell Physiol. 2010. PMID: 19847803 Review.
Cited by
-
Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells.Exp Hematol. 2010 May;38(5):403-16. doi: 10.1016/j.exphem.2010.02.011. Epub 2010 Mar 3. Exp Hematol. 2010. PMID: 20206228 Free PMC article.
-
Gene product diversity: adaptive or not?Trends Genet. 2022 Nov;38(11):1112-1122. doi: 10.1016/j.tig.2022.05.002. Epub 2022 May 28. Trends Genet. 2022. PMID: 35641344 Free PMC article. Review.
-
High-resolution analysis of cell-state transitions in yeast suggests widespread transcriptional tuning by alternative starts.Genome Biol. 2021 Jan 14;22(1):34. doi: 10.1186/s13059-020-02245-3. Genome Biol. 2021. PMID: 33446241 Free PMC article.
-
Central Role of Core Binding Factor β2 in Mucosa-Associated Lymphoid Tissue Organogenesis in Mouse.PLoS One. 2015 May 22;10(5):e0127460. doi: 10.1371/journal.pone.0127460. eCollection 2015. PLoS One. 2015. PMID: 26001080 Free PMC article.
-
Hematopoietic stem cell emergence in the conceptus and the role of Runx1.Int J Dev Biol. 2010;54(6-7):1151-63. doi: 10.1387/ijdb.103106gs. Int J Dev Biol. 2010. PMID: 20711992 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

