Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells
- PMID: 17626080
- PMCID: PMC2045433
- DOI: 10.1128/JVI.01009-07
Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells
Abstract
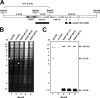
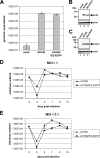
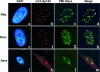
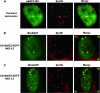
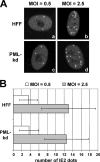
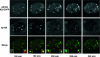
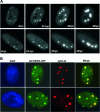
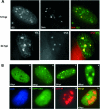
The human cytomegalovirus (HCMV) immediate-early 2 (IE2) transactivator has previously been shown to form intranuclear, dot-like accumulations in association with subnuclear structures known as promyelocytic leukemia protein (PML) nuclear bodies or ND10. We recently observed that IE2 can form dot-like structures even after infection of PML knockdown cells, which lack genuine ND10. To further analyze the determinants of IE2 subnuclear localization, a recombinant HCMV expressing IE2 fused to the enhanced green fluorescent protein was constructed. We infected primary human fibroblasts expressing Sp100 fused to the autofluorescent protein mCherry while performing live-cell imaging experiments. These experiments revealed a very dynamic association of IE2 dots with ND10 structures during the first hours postinfection: juxtaposed structures rapidly fused to precise co-localizations, followed by segregation, and finally, the dispersal of ND10 accumulations. Furthermore, by infecting PML knockdown cells we determined that the number of IE2 accumulations was dependent on the multiplicity of infection. Since time-lapse microscopy in live-infected cells revealed that IE2 foci developed into viral replication compartments, we hypothesized that viral DNA could act as a determinant of IE2 accumulations. Direct evidence that IE2 molecules are associated with viral DNA early after HCMV infection was obtained using fluorescence in situ hybridization. Finally, a DNA-binding-deficient IE2 mutant could no longer be recruited into viral replication centers, suggesting that the association of IE2 with viral DNA is mediated by a direct DNA contact. Thus, we identified viral DNA as an important determinant of IE2 subnuclear localization, which suggests that the formation of a virus-induced nucleoprotein complex and its spatial organization is likely to be critical at the early stages of a lytic infection.
Figures

Similar articles
-
Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections.J Virol. 2006 Aug;80(16):8006-18. doi: 10.1128/JVI.00743-06. J Virol. 2006. PMID: 16873257 Free PMC article.
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10).J Virol. 1999 Dec;73(12):10458-71. doi: 10.1128/JVI.73.12.10458-10471.1999. J Virol. 1999. PMID: 10559364 Free PMC article.
-
Are promyelocytic leukaemia protein nuclear bodies a scaffold for caspase-2 programmed cell death?Trends Biochem Sci. 2007 Sep;32(9):400-6. doi: 10.1016/j.tibs.2007.08.001. Epub 2007 Aug 10. Trends Biochem Sci. 2007. PMID: 17693089 Review.
-
Nuclear dots: actors on many stages.Immunobiology. 1997 Dec;198(1-3):307-31. doi: 10.1016/S0171-2985(97)80051-4. Immunobiology. 1997. PMID: 9442402 Review.
Cited by
-
Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection.J Virol. 2009 Dec;83(24):12881-94. doi: 10.1128/JVI.01525-09. Epub 2009 Oct 7. J Virol. 2009. PMID: 19812159 Free PMC article.
-
The Role of Nuclear Antiviral Factors against Invading DNA Viruses: The Immediate Fate of Incoming Viral Genomes.Viruses. 2016 Oct 22;8(10):290. doi: 10.3390/v8100290. Viruses. 2016. PMID: 27782081 Free PMC article. Review.
-
Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation.J Gen Virol. 2010 Nov;91(Pt 11):2664-76. doi: 10.1099/vir.0.022301-0. Epub 2010 Jul 14. J Gen Virol. 2010. PMID: 20631086 Free PMC article.
-
Human Cytomegalovirus IE2 Both Activates and Represses Initiation and Modulates Elongation in a Context-Dependent Manner.mBio. 2022 Jun 28;13(3):e0033722. doi: 10.1128/mbio.00337-22. Epub 2022 May 17. mBio. 2022. PMID: 35579393 Free PMC article.
-
Differences between mouse and human cytomegalovirus interactions with their respective hosts at immediate early times of the replication cycle.Med Microbiol Immunol. 2008 Jun;197(2):241-9. doi: 10.1007/s00430-008-0078-1. Epub 2008 Feb 9. Med Microbiol Immunol. 2008. PMID: 18264718 Review.
References
-
- Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13:111-121. - PubMed
-
- Ahn, J.-H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. - PMC - PubMed
-
- Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. - PMC - PubMed
-
- Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

