Epstein-Barr virus exploits BSAP/Pax5 to achieve the B-cell specificity of its growth-transforming program
- PMID: 17626071
- PMCID: PMC2045388
- DOI: 10.1128/JVI.00358-07
Epstein-Barr virus exploits BSAP/Pax5 to achieve the B-cell specificity of its growth-transforming program
Abstract
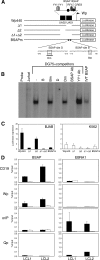
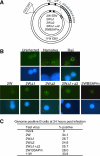
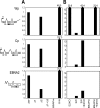
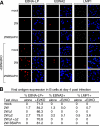
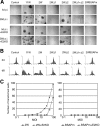
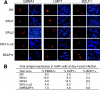
Epstein-Barr virus (EBV) can infect various cell types but limits its classical growth-transforming function to B lymphocytes, the cells in which it persists in vivo. Transformation initiates with the activation of Wp, a promoter present as tandemly repeated copies in the viral genome. Assays with short Wp reporter constructs have identified two promoter-activating regions, one of which (UAS2) appears to be lineage independent, while the other (UAS1) was B-cell specific and contained two putative binding sites for the B-cell-specific activator protein BSAP/Pax5. To address the physiologic relevance of these findings, we first used chromosome immunoprecipitation assays and found that BSAP is indeed bound to Wp sequences on the EBV genome in transformed cells. Thereafter, we constructed recombinant EBVs carrying two Wp copies, both wild type, with UAS1 or UAS2 deleted, or mutated in the BSAP binding sites. All the viruses delivered their genomes to the B-cell nucleus equally well. However, the BSAP binding mutant (and the virus with UAS1 deleted) showed no detectable activity in B cells, whether measured by early Wp transcription, expression of EBV latent proteins, or outgrowth of transformed cells. This was a B-cell-specific defect since, on entry into epithelial cells, an environment where Wp is not the latent promoter of choice, all the Wp mutant viruses initiated infection as efficiently as wild-type virus. We infer that EBV ensures the B-cell specificity of its growth-transforming function by exploiting BSAP/Pax5 as a lineage-specific activator of the transforming program.
Figures
Similar articles
-
The Epstein-Barr virus BamHI C promoter is not essential for B cell immortalization in vitro, but it greatly enhances B cell growth transformation.J Virol. 2015 Mar;89(5):2483-93. doi: 10.1128/JVI.03300-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540367 Free PMC article.
-
The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5.J Virol. 2000 Nov;74(22):10458-67. doi: 10.1128/jvi.74.22.10458-10467.2000. J Virol. 2000. PMID: 11044090 Free PMC article.
-
Characterisation of regulatory sequences at the Epstein-Barr virus BamHI W promoter.Virology. 1998 Dec 5;252(1):149-61. doi: 10.1006/viro.1998.9440. Virology. 1998. PMID: 9875325
-
Interaction of Epstein-Barr virus (EBV) with human B-lymphocytes.Biochem Biophys Res Commun. 2010 May 21;396(1):67-73. doi: 10.1016/j.bbrc.2010.02.146. Biochem Biophys Res Commun. 2010. PMID: 20494113 Review.
-
Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review.
Cited by
-
EBV latency types adopt alternative chromatin conformations.PLoS Pathog. 2011 Jul;7(7):e1002180. doi: 10.1371/journal.ppat.1002180. Epub 2011 Jul 28. PLoS Pathog. 2011. PMID: 21829357 Free PMC article.
-
The Epstein-Barr virus BamHI C promoter is not essential for B cell immortalization in vitro, but it greatly enhances B cell growth transformation.J Virol. 2015 Mar;89(5):2483-93. doi: 10.1128/JVI.03300-14. Epub 2014 Dec 24. J Virol. 2015. PMID: 25540367 Free PMC article.
-
Contributions of CTCF and DNA methyltransferases DNMT1 and DNMT3B to Epstein-Barr virus restricted latency.J Virol. 2012 Jan;86(2):1034-45. doi: 10.1128/JVI.05923-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072770 Free PMC article.
-
The B-cell-specific transcription factor and master regulator Pax5 promotes Epstein-Barr virus latency by negatively regulating the viral immediate early protein BZLF1.J Virol. 2013 Jul;87(14):8053-63. doi: 10.1128/JVI.00546-13. Epub 2013 May 15. J Virol. 2013. PMID: 23678172 Free PMC article.
-
Chromatin organization of gammaherpesvirus latent genomes.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):236-45. doi: 10.1016/j.bbagrm.2009.10.004. Epub 2009 Oct 22. Biochim Biophys Acta. 2010. PMID: 19853673 Free PMC article. Review.
References
-
- Adams, B., P. Dorfler, A. Aguzzi, Z. Kozmik, P. Urbanek, I. Maurer-Fogy, and M. Busslinger. 1992. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 6:1589-1607. - PubMed
-
- Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. - PubMed
-
- Allan, G. J., and D. T. Rowe. 1989. Size and stability of the Epstein-Barr virus major internal repeat (IR-1) in Burkitt's lymphoma and lymphoblastoid cell lines. Virology 173:489-498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

