Chd1 and yFACT act in opposition in regulating transcription
- PMID: 17620414
- PMCID: PMC2099615
- DOI: 10.1128/MCB.00978-07
Chd1 and yFACT act in opposition in regulating transcription
Abstract
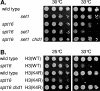
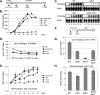
CHD1 encodes an ATP-dependent chromatin remodeler with two chromodomains. Deletion of CHD1 suppresses the temperature-sensitive growth defect caused by mutations in either SPT16 or POB3, which encode subunits of the yFACT chromatin-reorganizing complex. chd1 also suppresses synthetic defects caused by combining an spt16 mutation with other transcription factor mutations, including the synthetic lethality caused by combining an spt16 mutation with TATA binding protein (TBP) or TFIIA defects. Binding of TBP and RNA polymerase II to the GAL1 promoter is reduced in a pob3 mutant, resulting in low levels of GAL1 expression, and all three defects are suppressed by removing Chd1. These results suggest that Chd1 and yFACT have opposing roles in regulating TBP binding at promoters. Additionally, overexpression of Chd1 is tolerated in wild-type cells but is toxic in spt16 mutants. Further, both the ATPase and chromodomain are required for Chd1 activity in opposing yFACT function. Similar to the suppression by chd1, mutations in the SET2 histone methyltransferase also suppress defects caused by yFACT mutations. chd1 and set2 are additive in suppressing pob3, suggesting that Chd1 and Set2 act in distinct pathways. Although human Chd1 has been shown to bind to H3-K4-Me, we discuss evidence arguing that yeast Chd1 binds to neither H3-K4-Me nor H3-K36-Me.
Figures







Similar articles
-
Opposing roles for Set2 and yFACT in regulating TBP binding at promoters.EMBO J. 2006 Oct 4;25(19):4479-89. doi: 10.1038/sj.emboj.7601333. Epub 2006 Sep 14. EMBO J. 2006. PMID: 16977311 Free PMC article.
-
The yeast FACT complex has a role in transcriptional initiation.Mol Cell Biol. 2005 Jul;25(14):5812-22. doi: 10.1128/MCB.25.14.5812-5822.2005. Mol Cell Biol. 2005. PMID: 15987999 Free PMC article.
-
A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae.Genetics. 2008 Feb;178(2):649-59. doi: 10.1534/genetics.107.084202. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245327 Free PMC article.
-
Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article.
-
CHD1, a multifaceted epigenetic remodeler in prostate cancer.Front Oncol. 2023 Jan 26;13:1123362. doi: 10.3389/fonc.2023.1123362. eCollection 2023. Front Oncol. 2023. PMID: 36776288 Free PMC article. Review.
Cited by
-
Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription.PLoS Genet. 2009 Jan;5(1):e1000339. doi: 10.1371/journal.pgen.1000339. Epub 2009 Jan 16. PLoS Genet. 2009. PMID: 19148280 Free PMC article.
-
FACT interacts with Set3 HDAC and fine-tunes GAL1 transcription in response to environmental stimulation.Nucleic Acids Res. 2021 Jun 4;49(10):5502-5519. doi: 10.1093/nar/gkab312. Nucleic Acids Res. 2021. PMID: 33963860 Free PMC article.
-
Essential histone chaperones collaborate to regulate transcription and chromatin integrity.Genes Dev. 2021 May 1;35(9-10):698-712. doi: 10.1101/gad.348431.121. Epub 2021 Apr 22. Genes Dev. 2021. PMID: 33888559 Free PMC article.
-
Identification of Mutant Versions of the Spt16 Histone Chaperone That Are Defective for Transcription-Coupled Nucleosome Occupancy in Saccharomyces cerevisiae.G3 (Bethesda). 2012 May;2(5):555-67. doi: 10.1534/g3.112.002451. Epub 2012 May 1. G3 (Bethesda). 2012. PMID: 22670226 Free PMC article.
-
Comparative Genomics Reveals Chd1 as a Determinant of Nucleosome Spacing in Vivo.G3 (Bethesda). 2015 Jul 14;5(9):1889-97. doi: 10.1534/g3.115.020271. G3 (Bethesda). 2015. PMID: 26175451 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley and Sons, New York, NY.
-
- Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
